+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 spike trimer in complex with Fab NA8, ensemble map | |||||||||
 Map data Map data | Map after post-processing | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
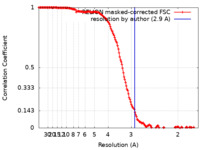
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Tsybovsky Y / Kwong PD / Farci P | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2022 Journal: Cell Rep / Year: 2022Title: Potent monoclonal antibodies neutralize Omicron sublineages and other SARS-CoV-2 variants. Authors: Zhaochun Chen / Peng Zhang / Yumiko Matsuoka / Yaroslav Tsybovsky / Kamille West / Celia Santos / Lisa F Boyd / Hanh Nguyen / Anna Pomerenke / Tyler Stephens / Adam S Olia / Baoshan Zhang / ...Authors: Zhaochun Chen / Peng Zhang / Yumiko Matsuoka / Yaroslav Tsybovsky / Kamille West / Celia Santos / Lisa F Boyd / Hanh Nguyen / Anna Pomerenke / Tyler Stephens / Adam S Olia / Baoshan Zhang / Valeria De Giorgi / Michael R Holbrook / Robin Gross / Elena Postnikova / Nicole L Garza / Reed F Johnson / David H Margulies / Peter D Kwong / Harvey J Alter / Ursula J Buchholz / Paolo Lusso / Patrizia Farci /  Abstract: The emergence and global spread of the SARS-CoV-2 Omicron variants, which carry an unprecedented number of mutations, raise serious concerns due to the reduced efficacy of current vaccines and ...The emergence and global spread of the SARS-CoV-2 Omicron variants, which carry an unprecedented number of mutations, raise serious concerns due to the reduced efficacy of current vaccines and resistance to therapeutic antibodies. Here, we report the generation and characterization of two potent human monoclonal antibodies, NA8 and NE12, against the receptor-binding domain of the SARS-CoV-2 spike protein. NA8 interacts with a highly conserved region and has a breadth of neutralization with picomolar potency against the Beta variant and the Omicron BA.1 and BA.2 sublineages and nanomolar potency against BA.2.12.1 and BA.4. Combination of NA8 and NE12 retains potent neutralizing activity against the major SARS-CoV-2 variants of concern. Cryo-EM analysis provides the structural basis for the broad and complementary neutralizing activity of these two antibodies. We confirm the in vivo protective and therapeutic efficacies of NA8 and NE12 in the hamster model. These results show that broad and potent human antibodies can overcome the continuous immune escape of evolving SARS-CoV-2 variants. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26403.map.gz emd_26403.map.gz | 24.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26403-v30.xml emd-26403-v30.xml emd-26403.xml emd-26403.xml | 21.6 KB 21.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26403_fsc.xml emd_26403_fsc.xml | 15.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_26403.png emd_26403.png | 60.4 KB | ||
| Masks |  emd_26403_msk_1.map emd_26403_msk_1.map | 325 MB |  Mask map Mask map | |
| Others |  emd_26403_additional_1.map.gz emd_26403_additional_1.map.gz emd_26403_half_map_1.map.gz emd_26403_half_map_1.map.gz emd_26403_half_map_2.map.gz emd_26403_half_map_2.map.gz | 260.2 MB 261.4 MB 261.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26403 http://ftp.pdbj.org/pub/emdb/structures/EMD-26403 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26403 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26403 | HTTPS FTP |
-Validation report
| Summary document |  emd_26403_validation.pdf.gz emd_26403_validation.pdf.gz | 680.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26403_full_validation.pdf.gz emd_26403_full_validation.pdf.gz | 679.7 KB | Display | |
| Data in XML |  emd_26403_validation.xml.gz emd_26403_validation.xml.gz | 23.2 KB | Display | |
| Data in CIF |  emd_26403_validation.cif.gz emd_26403_validation.cif.gz | 30.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26403 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26403 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26403 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26403 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26403.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26403.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map after post-processing | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.873 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26403_msk_1.map emd_26403_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Map before post-processing
| File | emd_26403_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map before post-processing | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 1
| File | emd_26403_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 2
| File | emd_26403_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 S6P spike trimer in complex with Fab NA8
| Entire | Name: SARS-CoV-2 S6P spike trimer in complex with Fab NA8 |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 S6P spike trimer in complex with Fab NA8
| Supramolecule | Name: SARS-CoV-2 S6P spike trimer in complex with Fab NA8 / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 580 KDa |
-Macromolecule #1: SARS-CoV-2 S6P spike trimer
| Macromolecule | Name: SARS-CoV-2 S6P spike trimer / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QCVNLTTRTQ LPPAYTNSFT RGVYYPDKVF RSSVLHSTQD LFLPF FSNV TWFHAIHVSG TNGTKRFDNP VLPFNDGVYF ASTEKSNIIR GWIFGT TLD SKTQSLLIVN NATNVVIKVC EFQFCNDPFL GVYYHKNNKS WMESEFR VY SSANNCTFEY VSQPFLMDLE ...String: QCVNLTTRTQ LPPAYTNSFT RGVYYPDKVF RSSVLHSTQD LFLPF FSNV TWFHAIHVSG TNGTKRFDNP VLPFNDGVYF ASTEKSNIIR GWIFGT TLD SKTQSLLIVN NATNVVIKVC EFQFCNDPFL GVYYHKNNKS WMESEFR VY SSANNCTFEY VSQPFLMDLE GKQGNFKNLR EFVFKNIDGY FKIYSKHT P INLVRDLPQG FSALEPLVDL PIGINITRFQ TLLALHRSYL TPGDSSSGW TAGAAAYYVG YLQPRTFLLK YNENGTITDA VDCALDPLSE TKCTLKSFTV EKGIYQTSN FRVQPTESIV RFPNITNLCP FGEVFNATRF ASVYAWNRKR I SNCVADYS VLYNSASFST FKCYGVSPTK LNDLCFTNVY ADSFVIRGDE VR QIAPGQT GKIADYNYKL PDDFTGCVIA WNSNNLDSKV GGNYNYLYRL FRK SNLKPF ERDISTEIYQ AGSTPCNGVE GFNCYFPLQS YGFQPTNGVG YQPY RVVVL SFELLHAPAT VCGPKKSTNL VKNKCVNFNF NGLTGTGVLT ESNKK FLPF QQFGRDIADT TDAVRDPQTL EILDITPCSF GGVSVITPGT NTSNQV AVL YQDVNCTEVP VAIHADQLTP TWRVYSTGSN VFQTRAGCLI GAEHVNN SY ECDIPIGAGI CASYQTQTNS PGSASSVASQ SIIAYTMSLG AENSVAYS N NSIAIPTNFT ISVTTEILPV SMTKTSVDCT MYICGDSTEC SNLLLQYGS FCTQLNRALT GIAVEQDKNT QEVFAQVKQI YKTPPIKDFG GFNFSQILPD PSKPSKRSP IEDLLFNKVT LADAGFIKQY GDCLGDIAAR DLICAQKFNG L TVLPPLLT DEMIAQYTSA LLAGTITSGW TFGAGPALQI PFPMQMAYRF NG IGVTQNV LYENQKLIAN QFNSAIGKIQ DSLSSTPSAL GKLQDVVNQN AQA LNTLVK QLSSNFGAIS SVLNDILSRL DPPEAEVQID RLITGRLQSL QTYV TQQLI RAAEIRASAN LAATKMSECV LGQSKRVDFC GKGYHLMSFP QSAPH GVVF LHVTYVPAQE KNFTTAPAIC HDGKAHFPRE GVFVSNGTHW FVTQRN FYE PQIITTDNTF VSGNCDVVIG IVNNTVYDPL QPELDSFKEE LDKYFKN HT SPDVDLGDIS GINASVVNIQ KEIDRLNEVA KNLNESLIDL QELGKYEQ G SGYIPEAPRD GQAYVRKDGE WVLLSTFLGR SLEVLFQGPG HHHHHHHHS AWSHPQFEKG GGSGGGGSGG SAWSHPQFEK |
-Macromolecule #2: NA8 Fab heavy chain
| Macromolecule | Name: NA8 Fab heavy chain / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: VQLLEESGGG AVQPGRSLRL SCEASGFSFN SYGMHWVRQA PGKGLEWVAA IWYSGSDRDY ADSVKGRFS ISRDNSKNTL YLQMNSLRAE DTAVYYCARD PHCTGGVCDA FDLWGQGTMV T VSSASTKG PSVFPGAPSS KSTSGGTAAL GCLVKDYFPE PVTVSWNSGA ...String: VQLLEESGGG AVQPGRSLRL SCEASGFSFN SYGMHWVRQA PGKGLEWVAA IWYSGSDRDY ADSVKGRFS ISRDNSKNTL YLQMNSLRAE DTAVYYCARD PHCTGGVCDA FDLWGQGTMV T VSSASTKG PSVFPGAPSS KSTSGGTAAL GCLVKDYFPE PVTVSWNSGA LTSGVHTFPA VL QSSGLYS LSSVVTVPSS SLGTQTYICN VDHKPATPRW TRKLSPNLVT KL |
-Macromolecule #3: NA8 Fab light chain
| Macromolecule | Name: NA8 Fab light chain / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: ELQMTQSPSS LSASVGDRVT ITCRASQSIS SYLNWYQQKP GKAPKLLIYA ASSLQSGVPS RFSGSGSGT DFTLTISSLQ PEDFATYYCQ QSYSTPYTFG QGTKLEIKRT VAAPSVFIFP P SDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN SQESVTEQDS ...String: ELQMTQSPSS LSASVGDRVT ITCRASQSIS SYLNWYQQKP GKAPKLLIYA ASSLQSGVPS RFSGSGSGT DFTLTISSLQ PEDFATYYCQ QSYSTPYTFG QGTKLEIKRT VAAPSVFIFP P SDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN SQESVTEQDS KDSTYSLSST LT LSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: PBS |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)