+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rabies virus glycoprotein trimer | |||||||||||||||
 Map data Map data | RABVG pre-fusion trimer with no interacting fusion loops | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Rabies virus glycoprotein / antibodies / viral fusion protein / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||||||||
| Biological species |  Rabies virus / Rabies virus /  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.92 Å | |||||||||||||||
 Authors Authors | Callaway HM / Zyla D / Larrous F / Dias de Melo G / Hastie KM / Avalos RD / Agarwal A / Bouhry H / Corti D / Saphire EO | |||||||||||||||
| Funding support |  United States, United States,  Switzerland, 4 items Switzerland, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Structure of the rabies virus glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Authors: Heather M Callaway / Dawid Zyla / Florence Larrous / Guilherme Dias de Melo / Kathryn M Hastie / Ruben Diaz Avalos / Alyssa Agarwal / Davide Corti / Hervé Bourhy / Erica Ollmann Saphire /    Abstract: Rabies infection is nearly 100% lethal if untreated and kills more than 50,000 people annually, many of them children. Existing rabies vaccines target the rabies virus glycoprotein (RABV-G) but ...Rabies infection is nearly 100% lethal if untreated and kills more than 50,000 people annually, many of them children. Existing rabies vaccines target the rabies virus glycoprotein (RABV-G) but generate short-lived immune responses, likely because the protein is heterogeneous under physiological conditions. Here, we report the 3.39 Å cryo-electron microscopy structure of trimeric, prefusion RABV-G complexed with RVA122, a potently neutralizing human antibody. RVA122 binds to a quaternary epitope at the top of RABV-G, bridging domains and stabilizing RABV-G protomers in a prefusion state. RABV-G trimerization involves side-to-side interactions between the central α helix and adjacent loops, rather than contacts between central helices, and interactions among the fusion loops at the glycoprotein base. These results provide a basis from which to develop improved rabies vaccines based on RABV-G stabilized in the prefusion conformation. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26398.map.gz emd_26398.map.gz | 2.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26398-v30.xml emd-26398-v30.xml emd-26398.xml emd-26398.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26398.png emd_26398.png | 472.8 KB | ||
| Filedesc metadata |  emd-26398.cif.gz emd-26398.cif.gz | 5.8 KB | ||
| Others |  emd_26398_half_map_1.map.gz emd_26398_half_map_1.map.gz emd_26398_half_map_2.map.gz emd_26398_half_map_2.map.gz | 4.7 MB 4.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26398 http://ftp.pdbj.org/pub/emdb/structures/EMD-26398 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26398 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26398 | HTTPS FTP |
-Validation report
| Summary document |  emd_26398_validation.pdf.gz emd_26398_validation.pdf.gz | 798.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26398_full_validation.pdf.gz emd_26398_full_validation.pdf.gz | 798.4 KB | Display | |
| Data in XML |  emd_26398_validation.xml.gz emd_26398_validation.xml.gz | 8.4 KB | Display | |
| Data in CIF |  emd_26398_validation.cif.gz emd_26398_validation.cif.gz | 9.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26398 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26398 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26398 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26398 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26398.map.gz / Format: CCP4 / Size: 5.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26398.map.gz / Format: CCP4 / Size: 5.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RABVG pre-fusion trimer with no interacting fusion loops | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.67636 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Map Half 1
| File | emd_26398_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map Half 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
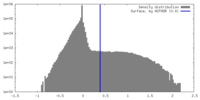
| Density Histograms |
-Half map: Hap Half 2
| File | emd_26398_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Hap Half 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
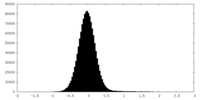
| Density Histograms |
- Sample components
Sample components
-Entire : Rabies virus glycoprotein pre-fusion trimer in complex with neutr...
| Entire | Name: Rabies virus glycoprotein pre-fusion trimer in complex with neutralizing antibody RVA122 |
|---|---|
| Components |
|
-Supramolecule #1: Rabies virus glycoprotein pre-fusion trimer in complex with neutr...
| Supramolecule | Name: Rabies virus glycoprotein pre-fusion trimer in complex with neutralizing antibody RVA122 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Reconstruction with no interacting fusion loops |
|---|
-Macromolecule #1: Glycoprotein
| Macromolecule | Name: Glycoprotein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rabies virus / Strain: PV Rabies virus / Strain: PV |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MVPQALLFVP LLVFPLCFGK FPIYTIPDKL GPWSPIDIHH LSCPNNLVVE DEGCTNLSGF SYMELKVGYI SAIKMNGFTC TGVVTEAET YTNFVGYVTT TFKRKHFRPT PDACRAAYNW KMAGDPRYEE SLHNPYPDYH WLRTVKTTKE SLVIISPSVA D LDPYDRSL ...String: MVPQALLFVP LLVFPLCFGK FPIYTIPDKL GPWSPIDIHH LSCPNNLVVE DEGCTNLSGF SYMELKVGYI SAIKMNGFTC TGVVTEAET YTNFVGYVTT TFKRKHFRPT PDACRAAYNW KMAGDPRYEE SLHNPYPDYH WLRTVKTTKE SLVIISPSVA D LDPYDRSL HSRVFPGGNC SGVAVSSTYC STNHDYTIWM PENPRLGMSC DIFTNSRGKR ASKGSETCGF VDERGLYKSL KG ACKLKLC GVLGLRLMDG TWVAMQTSNE TKWCPPGQLV NLHDFRSDEI EHLVVEELVK KREECLDALE SIMTTKSVSF RRL SHLRKL VPGFGKAYTI FNKTLMEADA HYKSVRTWNE IIPSKGCLRV GGRCHPHVNG VFFNGIILGP DGNVLIPEMQ SSLL QQHME LLVSSVIPLM HPLADPSTVF KNGDEAEDFV EVHL |
-Macromolecule #2: RVA122 Fab Light Chain
| Macromolecule | Name: RVA122 Fab Light Chain / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: QSVLTQSPSA SDTPGQRVTI SCSGSSSNIG SNYVYWYQQF PGTAPKLLIY KSDKRPSGVP DRFSGSTSGT SASLAISGLR SEDEADYYC AAWDNRLSGW LFGGGTKLTV LGTVAAPSVF IFPPSDEQLK SGTASVVCLL NNFYPREAKV QWKVDNALQS G NSQESVTE ...String: QSVLTQSPSA SDTPGQRVTI SCSGSSSNIG SNYVYWYQQF PGTAPKLLIY KSDKRPSGVP DRFSGSTSGT SASLAISGLR SEDEADYYC AAWDNRLSGW LFGGGTKLTV LGTVAAPSVF IFPPSDEQLK SGTASVVCLL NNFYPREAKV QWKVDNALQS G NSQESVTE QDSKDSTYSL SSTLTLSKAD YEKHKVYACE VTHQGLRSPV TKSFNRGEC |
-Macromolecule #3: RVA122 Fab Heavy Chain
| Macromolecule | Name: RVA122 Fab Heavy Chain / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: QVHLQESGPG LVKPSETLSL TCTVSGDSMN NFYWGWIRQP AGKGLEWIGY IYYSGTTNYN PSLKSRVTIS IDTSKNQFSL KVNSVTAAD TAVYYCARDS GDYVSYYYYG MDVWGPGTTV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA ...String: QVHLQESGPG LVKPSETLSL TCTVSGDSMN NFYWGWIRQP AGKGLEWIGY IYYSGTTNYN PSLKSRVTIS IDTSKNQFSL KVNSVTAAD TAVYYCARDS GDYVSYYYYG MDVWGPGTTV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYICNV NHKPSNTKVD KKVEPKSCDL EVDDDDKAGW SH PQFEKGG GSGGGSGGGS WSHPQFEK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: PBS pH 7.4 with 0.03mM LMNG | ||||||||||||||||||
| Grid | Model: C-flat-1.2/1.3 | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 BASE (4k x 4k) / Average electron dose: 26.74 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)