+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | HIV-1 Envelope ApexGT2 in complex with RM20A3 Fab | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | HIV / germline targeting vaccine / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
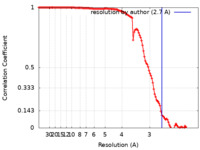
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||||||||
 Authors Authors | Berndsen ZT / Ward AB | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Immunity / Year: 2022 Journal: Immunity / Year: 2022Title: Human immunoglobulin repertoire analysis guides design of vaccine priming immunogens targeting HIV V2-apex broadly neutralizing antibody precursors. Authors: Jordan R Willis / Zachary T Berndsen / Krystal M Ma / Jon M Steichen / Torben Schiffner / Elise Landais / Alessia Liguori / Oleksandr Kalyuzhniy / Joel D Allen / Sabyasachi Baboo / ...Authors: Jordan R Willis / Zachary T Berndsen / Krystal M Ma / Jon M Steichen / Torben Schiffner / Elise Landais / Alessia Liguori / Oleksandr Kalyuzhniy / Joel D Allen / Sabyasachi Baboo / Oluwarotimi Omorodion / Jolene K Diedrich / Xiaozhen Hu / Erik Georgeson / Nicole Phelps / Saman Eskandarzadeh / Bettina Groschel / Michael Kubitz / Yumiko Adachi / Tina-Marie Mullin / Nushin B Alavi / Samantha Falcone / Sunny Himansu / Andrea Carfi / Ian A Wilson / John R Yates / James C Paulson / Max Crispin / Andrew B Ward / William R Schief /   Abstract: Broadly neutralizing antibodies (bnAbs) to the HIV envelope (Env) V2-apex region are important leads for HIV vaccine design. Most V2-apex bnAbs engage Env with an uncommonly long heavy-chain ...Broadly neutralizing antibodies (bnAbs) to the HIV envelope (Env) V2-apex region are important leads for HIV vaccine design. Most V2-apex bnAbs engage Env with an uncommonly long heavy-chain complementarity-determining region 3 (HCDR3), suggesting that the rarity of bnAb precursors poses a challenge for vaccine priming. We created precursor sequence definitions for V2-apex HCDR3-dependent bnAbs and searched for related precursors in human antibody heavy-chain ultradeep sequencing data from 14 HIV-unexposed donors. We found potential precursors in a majority of donors for only two long-HCDR3 V2-apex bnAbs, PCT64 and PG9, identifying these bnAbs as priority vaccine targets. We then engineered ApexGT Env trimers that bound inferred germlines for PCT64 and PG9 and had higher affinities for bnAbs, determined cryo-EM structures of ApexGT trimers complexed with inferred-germline and bnAb forms of PCT64 and PG9, and developed an mRNA-encoded cell-surface ApexGT trimer. These methods and immunogens have promise to assist HIV vaccine development. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25734.map.gz emd_25734.map.gz | 154.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25734-v30.xml emd-25734-v30.xml emd-25734.xml emd-25734.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25734_fsc.xml emd_25734_fsc.xml | 12.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_25734.png emd_25734.png | 131 KB | ||
| Masks |  emd_25734_msk_1.map emd_25734_msk_1.map | 163.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-25734.cif.gz emd-25734.cif.gz | 6.5 KB | ||
| Others |  emd_25734_half_map_1.map.gz emd_25734_half_map_1.map.gz emd_25734_half_map_2.map.gz emd_25734_half_map_2.map.gz | 151.7 MB 151.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25734 http://ftp.pdbj.org/pub/emdb/structures/EMD-25734 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25734 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25734 | HTTPS FTP |
-Validation report
| Summary document |  emd_25734_validation.pdf.gz emd_25734_validation.pdf.gz | 864 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25734_full_validation.pdf.gz emd_25734_full_validation.pdf.gz | 863.6 KB | Display | |
| Data in XML |  emd_25734_validation.xml.gz emd_25734_validation.xml.gz | 20.1 KB | Display | |
| Data in CIF |  emd_25734_validation.cif.gz emd_25734_validation.cif.gz | 26.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25734 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25734 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25734 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25734 | HTTPS FTP |
-Related structure data
| Related structure data |  7t75MC  7t73C  7t74C  7t76C  7t77C  7ti6C C: citing same article ( M: atomic model generated by this map |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25734.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25734.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||
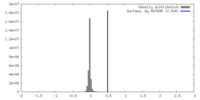
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_25734_msk_1.map emd_25734_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
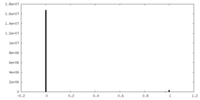
| Density Histograms |
-Half map: #2
| File | emd_25734_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
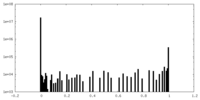
| Density Histograms |
-Half map: #1
| File | emd_25734_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
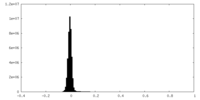
| Density Histograms |
- Sample components
Sample components
-Entire : HIV-1 Envelope ApexGT2 in complex with RM20A3 Fab
| Entire | Name: HIV-1 Envelope ApexGT2 in complex with RM20A3 Fab |
|---|---|
| Components |
|
-Supramolecule #1: HIV-1 Envelope ApexGT2 in complex with RM20A3 Fab
| Supramolecule | Name: HIV-1 Envelope ApexGT2 in complex with RM20A3 Fab / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: HIV Envelope ApexGT2 gp120
| Macromolecule | Name: HIV Envelope ApexGT2 gp120 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 56.12302 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGILPSPGMP ALLSLVSLLS VLLMGCVAET GAENLWVTVY YGVPVWKDAE TTLFCASDAK AYETEKHNVW ATHACVPTDP NPQEIHLEN VTEEFNMWKN NMVEQMHEDI ISLWDQSLKP CVKLTPLCVT LQCTNVTNNI TDDMRGELKN CSFNATTELR N KRQKVYSL ...String: MGILPSPGMP ALLSLVSLLS VLLMGCVAET GAENLWVTVY YGVPVWKDAE TTLFCASDAK AYETEKHNVW ATHACVPTDP NPQEIHLEN VTEEFNMWKN NMVEQMHEDI ISLWDQSLKP CVKLTPLCVT LQCTNVTNNI TDDMRGELKN CSFNATTELR N KRQKVYSL FYRLDIVPMG ENSTNYRLIN CNTSAITQAC PKVSFEPIPI HYCAPAGFAI LKCKDKKFNG TGPCPSVSTV QC THGIKPV VSTQLLLNGS LAEEEVIIRS ENITNNAKNI LVQLNTPVQI NCTRPNNNTV KSIRIGPGQA FYYTGDIIGD IRQ AHCNVS KATWNETLGK VVKQLRKHFG NNTIIRFAQS SGGDLEVTTH SFNCGGEFFY CNTSGLFNST WISNTSVQGS NSTG SNDSI TLPCRIKQII NMWQRIGQAM YAPPIQGVIR CVSNITGLIL TRDGGSTNST TETFRPGGGD MRDNWRSELY KYKVV KIEP LGVAPTRCKR RVVGRRRRRR |
-Macromolecule #2: HIV Envelope ApexGT2 gp41
| Macromolecule | Name: HIV Envelope ApexGT2 gp41 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 18.250541 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVSLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEP QQHLLKDTHW GIKQLQARVL AVEHYLRDQQ LLGIWGCSG KLICCTNVPW NSSWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALDGTKHHH H HH |
-Macromolecule #3: RM20A3 Fab Heavy Chain
| Macromolecule | Name: RM20A3 Fab Heavy Chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.511111 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVETGGG LVQPGGSLKL SCRASGYTFS SFAMSWVRQA PGKGLEWVSL INDRGGLTFY VDSVKGRFTI SRDNSKNTLS LQMHSLRDG DTAVYYCATG GMSSALQSSK YYFDFWGQGA LVTVSS |
-Macromolecule #4: RM20A3 Fab Light Chain
| Macromolecule | Name: RM20A3 Fab Light Chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.5088 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ALTQPPSVSG SPGQSVTISC TGTSSDIGSY NYVSWYQQHP GKAPKLMIYD VTQRPSGVSD RFSGSKSGNT ASLTISGLQA DDEADYYCS AYAGRQTFYI FGGGTRLTVL GQPKASPTVT LFPPSSEEL |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 15 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)