+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24643 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 Spike bound to Fab PDI 96 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Pymm P / Glukhova A / Black K / Tham WH | |||||||||
| Funding support |  Australia, 1 items Australia, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2021 Journal: Cell Rep / Year: 2021Title: Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain. Authors: Adam K Wheatley / Phillip Pymm / Robyn Esterbauer / Melanie H Dietrich / Wen Shi Lee / Damien Drew / Hannah G Kelly / Li-Jin Chan / Francesca L Mordant / Katrina A Black / Amy Adair / Hyon- ...Authors: Adam K Wheatley / Phillip Pymm / Robyn Esterbauer / Melanie H Dietrich / Wen Shi Lee / Damien Drew / Hannah G Kelly / Li-Jin Chan / Francesca L Mordant / Katrina A Black / Amy Adair / Hyon-Xhi Tan / Jennifer A Juno / Kathleen M Wragg / Thakshila Amarasena / Ester Lopez / Kevin J Selva / Ebene R Haycroft / James P Cooney / Hariprasad Venugopal / Li Lynn Tan / Matthew T O Neill / Cody C Allison / Deborah Cromer / Miles P Davenport / Richard A Bowen / Amy W Chung / Marc Pellegrini / Mark T Liddament / Alisa Glukhova / Kanta Subbarao / Stephen J Kent / Wai-Hong Tham /   Abstract: Potent neutralizing monoclonal antibodies are one of the few agents currently available to treat COVID-19. SARS-CoV-2 variants of concern (VOCs) that carry multiple mutations in the viral spike ...Potent neutralizing monoclonal antibodies are one of the few agents currently available to treat COVID-19. SARS-CoV-2 variants of concern (VOCs) that carry multiple mutations in the viral spike protein can exhibit neutralization resistance, potentially affecting the effectiveness of some antibody-based therapeutics. Here, the generation of a diverse panel of 91 human, neutralizing monoclonal antibodies provides an in-depth structural and phenotypic definition of receptor binding domain (RBD) antigenic sites on the viral spike. These RBD antibodies ameliorate SARS-CoV-2 infection in mice and hamster models in a dose-dependent manner and in proportion to in vitro, neutralizing potency. Assessing the effect of mutations in the spike protein on antibody recognition and neutralization highlights both potent single antibodies and stereotypic classes of antibodies that are unaffected by currently circulating VOCs, such as B.1.351 and P.1. These neutralizing monoclonal antibodies and others that bind analogous epitopes represent potentially useful future anti-SARS-CoV-2 therapeutics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24643.map.gz emd_24643.map.gz | 3.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24643-v30.xml emd-24643-v30.xml emd-24643.xml emd-24643.xml | 11.2 KB 11.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24643_fsc.xml emd_24643_fsc.xml | 8 KB | Display |  FSC data file FSC data file |
| Images |  emd_24643.png emd_24643.png | 42.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24643 http://ftp.pdbj.org/pub/emdb/structures/EMD-24643 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24643 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24643 | HTTPS FTP |
-Validation report
| Summary document |  emd_24643_validation.pdf.gz emd_24643_validation.pdf.gz | 360.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24643_full_validation.pdf.gz emd_24643_full_validation.pdf.gz | 360.1 KB | Display | |
| Data in XML |  emd_24643_validation.xml.gz emd_24643_validation.xml.gz | 10.1 KB | Display | |
| Data in CIF |  emd_24643_validation.cif.gz emd_24643_validation.cif.gz | 13.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24643 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24643 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24643 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24643 | HTTPS FTP |
-Related structure data
| Related structure data |  7mzfC  7mzgC  7mzhC  7mziC  7mzjC  7mzkC  7mzlC  7mzmC  7mznC  7rr0C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24643.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24643.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
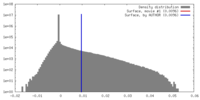
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : SARS-CoV-2 Spike- Fab PDI 96 complex
| Entire | Name: SARS-CoV-2 Spike- Fab PDI 96 complex |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 Spike- Fab PDI 96 complex
| Supramolecule | Name: SARS-CoV-2 Spike- Fab PDI 96 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Molecular weight | Theoretical: 560 KDa |
-Supramolecule #2: SARS-CoV-2 Spike
| Supramolecule | Name: SARS-CoV-2 Spike / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Fab PDI 96
| Supramolecule | Name: Fab PDI 96 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 80000 |
| Sample stage | Specimen holder model: OTHER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)