[English] 日本語
 Yorodumi
Yorodumi- EMDB-23861: Asymmetric structure of the uncleaved full-length HIV-1 envelope ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23861 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Asymmetric structure of the uncleaved full-length HIV-1 envelope glycoprotein trimer in state U2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane ...positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |   HIV-1 (virus) HIV-1 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.7 Å | |||||||||
 Authors Authors | Zhang SJ / Wang KY / Wang WL / Sodroksi J / Mao YD | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: J.Virol. / Year: 2021 Journal: J.Virol. / Year: 2021Title: Asymmetric Structures and Conformational Plasticity of the Uncleaved Full-Length Human Immunodeficiency Virus Envelope Glycoprotein Trimer. Authors: Zhang S / Wang K / Wang WL / Nguyen HT / Chen S / Lu M / Go EP / Ding H / Steinbock RT / Desaire H / Kappes JC / Sodroski J / Mao Y | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23861.map.gz emd_23861.map.gz | 131.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23861-v30.xml emd-23861-v30.xml emd-23861.xml emd-23861.xml | 9.5 KB 9.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23861.png emd_23861.png | 180.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23861 http://ftp.pdbj.org/pub/emdb/structures/EMD-23861 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23861 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23861 | HTTPS FTP |
-Validation report
| Summary document |  emd_23861_validation.pdf.gz emd_23861_validation.pdf.gz | 353.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23861_full_validation.pdf.gz emd_23861_full_validation.pdf.gz | 352.9 KB | Display | |
| Data in XML |  emd_23861_validation.xml.gz emd_23861_validation.xml.gz | 6.3 KB | Display | |
| Data in CIF |  emd_23861_validation.cif.gz emd_23861_validation.cif.gz | 7.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23861 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23861 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23861 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23861 | HTTPS FTP |
-Related structure data
| Related structure data | 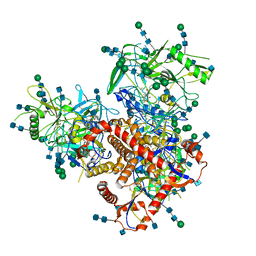 7n6wMC 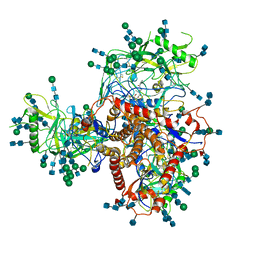 7n6uC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10163 (Title: Asymmetric structures of the uncleaved full-length HIV-1 envelope glycoprotein trimer EMPIAR-10163 (Title: Asymmetric structures of the uncleaved full-length HIV-1 envelope glycoprotein trimerData size: 4.0 TB Data #1: Drift-corrected micrographs of the uncleaved full-length HIV-1 envelope glycoprotein trimer (300 kV) [micrographs - single frame] Data #2: Drift-corrected micrographs of the uncleaved full-length HIV-1 envelope glycoprotein trimer (200 kV) [micrographs - single frame] Data #3: The classified particle stacks of the full-length HIV-1 envelope precursor in the P2 class [picked particles - single frame - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23861.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23861.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.685 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : The uncleaved full-length human immunodeficiency virus (HIV-1) en...
| Entire | Name: The uncleaved full-length human immunodeficiency virus (HIV-1) envelope glycoprotein trimer |
|---|---|
| Components |
|
-Supramolecule #1: The uncleaved full-length human immunodeficiency virus (HIV-1) en...
| Supramolecule | Name: The uncleaved full-length human immunodeficiency virus (HIV-1) envelope glycoprotein trimer type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   HIV-1 (virus) HIV-1 (virus) |
| Recombinant expression | Organism:  |
-Macromolecule #1: HIV-1 gp160 glycoprotein
| Macromolecule | Name: HIV-1 gp160 glycoprotein / type: protein_or_peptide / ID: 1 / Enantiomer: DEXTRO |
|---|---|
| Sequence | String: MRVKEKYQHL WRWGWRWGTM LLGMLMICSA TEKLWVTVYY GVPVWKEATT TLFCASDAKA YDTEVHNVWA THACVPTDPN PQEVVLENVT EHFNMWKNNM VEQMQEDIIS LWDQSLKPCV KLTPLCVTLN CKDVNATNTT NDSEGTMERG EIKNCSFNIT TSIRDEVQKE ...String: MRVKEKYQHL WRWGWRWGTM LLGMLMICSA TEKLWVTVYY GVPVWKEATT TLFCASDAKA YDTEVHNVWA THACVPTDPN PQEVVLENVT EHFNMWKNNM VEQMQEDIIS LWDQSLKPCV KLTPLCVTLN CKDVNATNTT NDSEGTMERG EIKNCSFNIT TSIRDEVQKE YALFYKLDVV PIDNNNTSYR LISCDTSVIT QACPKISFEP IPIHYCAPAG FAILKCNDKT FNGKGPCKNV STVQCTHGIR PVVSTQLLLN GSLAEEEVVI RSDNFTNNAK TIIVQLKESV EINCTRPNNN TRKSIHIGPG RAFYTTGEII GDIRQAHCNI SRAKWNDTLK QIVIKLREQF ENKTIVFNHS SGGDPEIVMH SFNCGGEFFY CNSTQLFNST WNNNTEGSNN TEGNTITLPC RIKQIINMWQ EVGKAMYAPP IRGQIRCSSN ITGLLLTRDG GINENGTEIF RPGGGDMRDN WRSELYKYKV VKIEPLGVAP TKAKRRVVQS EKSAVGIGAV FLGFLGAAGS TMGAASMTLT VQARLLLSGI VQQQNNLLRA IEAQQRMLQL TVWGIKQLQA RVLAVERYLG DQQLLGIWGC SGKLICTTAV PWNASWSNKS LDRIWNNMTW MEWEREIDNY TSEIYTLIEE SQNQQEKNEQ ELLELDKWAS LWNWFDITKW LWYIKIFIMI VGGLVGLRLV FTVLSIVNRV RQGYSPLSFQ TLLPAPRGPD RPEGIEEEGG ERDRDRSGRL VNGSLALIWD DLRSLCLFSY HRLRDLLLIV TRIVELLGRR GWEALKYWWN LLQYWSQELK NSAVSLLNAT AIAVAEGTDR VIEVVQGACR AIRHIPRRIR QGLERILL |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.7 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 55571 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)