[English] 日本語
 Yorodumi
Yorodumi- EMDB-23808: Structure of the phosphoinositide 3-kinase p110 gamma (PIK3CG) p1... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23808 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the phosphoinositide 3-kinase p110 gamma (PIK3CG) p101 (PIK3R5) complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PI3K / p110 / PIK3CG / PIK3R5 / p101 / phosphoinositide 3-kinase / PIP3 / IMMUNE SYSTEM / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of cardiac muscle contraction / negative regulation of triglyceride catabolic process / natural killer cell chemotaxis / secretory granule localization / neutrophil extravasation / phosphatidylinositol-4-phosphate 3-kinase / positive regulation of acute inflammatory response / 1-phosphatidylinositol-3-kinase regulator activity / respiratory burst involved in defense response / phosphatidylinositol 3-kinase complex ...negative regulation of cardiac muscle contraction / negative regulation of triglyceride catabolic process / natural killer cell chemotaxis / secretory granule localization / neutrophil extravasation / phosphatidylinositol-4-phosphate 3-kinase / positive regulation of acute inflammatory response / 1-phosphatidylinositol-3-kinase regulator activity / respiratory burst involved in defense response / phosphatidylinositol 3-kinase complex / T cell chemotaxis / negative regulation of fibroblast apoptotic process / phosphatidylinositol 3-kinase complex, class IB / regulation of calcium ion transmembrane transport / 1-phosphatidylinositol-4-phosphate 3-kinase activity / Co-stimulation by ICOS / sphingosine-1-phosphate receptor signaling pathway / phosphatidylinositol 3-kinase complex, class IA / phosphatidylinositol-3-phosphate biosynthetic process / 1-phosphatidylinositol-4,5-bisphosphate 3-kinase activity / phosphatidylinositol-4,5-bisphosphate 3-kinase / phosphatidylinositol 3-kinase / dendritic cell chemotaxis / 1-phosphatidylinositol-3-kinase activity / Erythropoietin activates Phosphoinositide-3-kinase (PI3K) / mast cell degranulation / hepatocyte apoptotic process / phosphatidylinositol-mediated signaling / regulation of cell adhesion mediated by integrin / phosphatidylinositol phosphate biosynthetic process / Synthesis of PIPs at the plasma membrane / positive regulation of MAP kinase activity / positive regulation of Rac protein signal transduction / CD28 dependent PI3K/Akt signaling / regulation of angiogenesis / T cell proliferation / ephrin receptor binding / GPVI-mediated activation cascade / neutrophil chemotaxis / positive regulation of endothelial cell migration / cellular response to cAMP / T cell activation / positive regulation of cytokine production / phosphatidylinositol 3-kinase/protein kinase B signal transduction / platelet aggregation / G-protein beta/gamma-subunit complex binding / endocytosis / Constitutive Signaling by Aberrant PI3K in Cancer / G beta:gamma signalling through PI3Kgamma / cell migration / PIP3 activates AKT signaling / positive regulation of cytosolic calcium ion concentration / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / angiogenesis / phospholipase C-activating G protein-coupled receptor signaling pathway / adaptive immune response / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / protein kinase activity / non-specific serine/threonine protein kinase / immune response / G protein-coupled receptor signaling pathway / inflammatory response / innate immune response / protein serine kinase activity / protein serine/threonine kinase activity / ATP binding / identical protein binding / nucleus / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
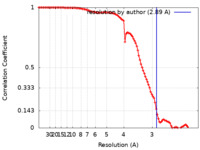
| Method | single particle reconstruction / cryo EM / Resolution: 2.89 Å | |||||||||
 Authors Authors | Burke JE / Dalwadi U / Rathinaswamy MK / Yip CK | |||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2021 Journal: Structure / Year: 2021Title: HDX-MS-optimized approach to characterize nanobodies as tools for biochemical and structural studies of class IB phosphoinositide 3-kinases. Authors: Manoj K Rathinaswamy / Kaelin D Fleming / Udit Dalwadi / Els Pardon / Noah J Harris / Calvin K Yip / Jan Steyaert / John E Burke /   Abstract: There is considerable interest in developing antibodies as modulators of signaling pathways. One of the most important signaling pathways in higher eukaryotes is the phosphoinositide 3-kinase (PI3K) ...There is considerable interest in developing antibodies as modulators of signaling pathways. One of the most important signaling pathways in higher eukaryotes is the phosphoinositide 3-kinase (PI3K) pathway, which plays fundamental roles in growth, metabolism, and immunity. The class IB PI3K, PI3Kγ, is a heterodimeric complex composed of a catalytic p110γ subunit bound to a p101 or p84 regulatory subunit. PI3Kγ is a critical component in multiple immune signaling processes and is dependent on activation by Ras and G protein-coupled receptors (GPCRs) to mediate its cellular roles. Here we describe the rapid and efficient characterization of multiple PI3Kγ binding single-chain camelid nanobodies using hydrogen-deuterium exchange (HDX) mass spectrometry (MS) for structural and biochemical studies. We identify nanobodies that stimulated lipid kinase activity, block Ras activation, and specifically inhibited p101-mediated GPCR activation. Overall, our work reveals insight into PI3Kγ regulation and identifies sites that may be exploited for therapeutic development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23808.map.gz emd_23808.map.gz | 82.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23808-v30.xml emd-23808-v30.xml emd-23808.xml emd-23808.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23808_fsc.xml emd_23808_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_23808.png emd_23808.png | 131 KB | ||
| Filedesc metadata |  emd-23808.cif.gz emd-23808.cif.gz | 8.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23808 http://ftp.pdbj.org/pub/emdb/structures/EMD-23808 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23808 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23808 | HTTPS FTP |
-Validation report
| Summary document |  emd_23808_validation.pdf.gz emd_23808_validation.pdf.gz | 514.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23808_full_validation.pdf.gz emd_23808_full_validation.pdf.gz | 513.8 KB | Display | |
| Data in XML |  emd_23808_validation.xml.gz emd_23808_validation.xml.gz | 12.1 KB | Display | |
| Data in CIF |  emd_23808_validation.cif.gz emd_23808_validation.cif.gz | 16 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23808 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23808 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23808 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23808 | HTTPS FTP |
-Related structure data
| Related structure data |  7mezMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23808.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23808.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Ternary complex of p110 gamma with p101 and a p101 binding nanobody
| Entire | Name: Ternary complex of p110 gamma with p101 and a p101 binding nanobody |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of p110 gamma with p101 and a p101 binding nanobody
| Supramolecule | Name: Ternary complex of p110 gamma with p101 and a p101 binding nanobody type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Nanobody binds to GBD domain, but was too fleixble to be built in the atomic model |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit ...
| Macromolecule | Name: Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: phosphatidylinositol 3-kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 126.627406 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MELENYKQPV VLREDNCRRR RRMKPRSAAA SLSSMELIPI EFVLPTSQRK CKSPETALLH VAGHGNVEQM KAQVWLRALE TSVAADFYH RLGPHHFLLL YQKKGQWYEI YDKYQVVQTL DCLRYWKATH RSPGQIHLVQ RHPPSEESQA FQRQLTALIG Y DVTDVSNV ...String: MELENYKQPV VLREDNCRRR RRMKPRSAAA SLSSMELIPI EFVLPTSQRK CKSPETALLH VAGHGNVEQM KAQVWLRALE TSVAADFYH RLGPHHFLLL YQKKGQWYEI YDKYQVVQTL DCLRYWKATH RSPGQIHLVQ RHPPSEESQA FQRQLTALIG Y DVTDVSNV HDDELEFTRR GLVTPRMAEV ASRDPKLYAM HPWVTSKPLP EYLWKKIANN CIFIVIHRST TSQTIKVSPD DT PGAILQS FFTKMAKKKS LMDIPESQSE QDFVLRVCGR DEYLVGETPI KNFQWVRHCL KNGEEIHVVL DTPPDPALDE VRK EEWPLV DDCTGVTGYH EQLTIHGKDH ESVFTVSLWD CDRKFRVKIR GIDIPVLPRN TDLTVFVEAN IQHGQQVLCQ RRTS PKPFT EEVLWNVWLE FSIKIKDLPK GALLNLQIYC GKAPALSSKA SAESPSSESK GKVQLLYYVN LLLIDHRFLL RRGEY VLHM WQISGKGEDQ GSFNADKLTS ATNPDKENSM SISILLDNYC HPIALPKHQP TPDPEGDRVR AEMPNQLRKQ LEAIIA TDP LNPLTAEDKE LLWHFRYESL KHPKAYPKLF SSVKWGQQEI VAKTYQLLAR REVWDQSALD VGLTMQLLDC NFSDENV RA IAVQKLESLE DDDVLHYLLQ LVQAVKFEPY HDSALARFLL KRGLRNKRIG HFLFWFLRSE IAQSRHYQQR FAVILEAY L RGCGTAMLHD FTQQVQVIEM LQKVTLDIKS LSAEKYDVSS QVISQLKQKL ENLQNSQLPE SFRVPYDPGL KAGALAIEK CKVMASKKKP LWLEFKCADP TALSNETIGI IFKHGDDLRQ DMLILQILRI MESIWETESL DLCLLPYGCI STGDKIGMIE IVKDATTIA KIQQSTVGNT GAFKDEVLNH WLKEKSPTEE KFQAAVERFV YSCAGYCVAT FVLGIGDRHN DNIMITETGN L FHIDFGHI LGNYKSFLGI NKERVPFVLT PDFLFVMGTS GKKTSPHFQK FQDICVKAYL ALRHHTNLLI ILFSMMLMTG MP QLTSKED IEYIRDALTV GKNEEDAKKY FLDQIEVCRD KGWTVQFNWF LHLVLGIKQG EKHSA UniProtKB: Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform |
-Macromolecule #2: Phosphoinositide 3-kinase regulatory subunit 5
| Macromolecule | Name: Phosphoinositide 3-kinase regulatory subunit 5 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 97.471805 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAGTMQPGAT TCTEDRIQHA LERCLHGLSL SRRSTSWSAG LCLNCWSLQE LVSRDPGHFL ILLEQILQKT REVQEKGTYD LLAPLALLF YSTVLCTPHF PPDSDLLLKA ARTYHRFLTW PVPYCSICQE LLTFIDAELK APGISYQRLV RAEQGLSTRS H RSSTVTVL ...String: GAGTMQPGAT TCTEDRIQHA LERCLHGLSL SRRSTSWSAG LCLNCWSLQE LVSRDPGHFL ILLEQILQKT REVQEKGTYD LLAPLALLF YSTVLCTPHF PPDSDLLLKA ARTYHRFLTW PVPYCSICQE LLTFIDAELK APGISYQRLV RAEQGLSTRS H RSSTVTVL LLNPVEVQAE FLDVADKLST PGPSPHSAYI TLLLHAFQAT FGAHCDLSGL HRRLQSKTLA ELEAIFTETA EA QELASGI GDAAEARQWL RTKLQAVGEK AGFPGVLDTA KPGKLRTIPI PVARCYTYSW NQDSFDILQE ILLKEQELLQ PEI LDDEED EDEEDEEEDL DADGHCAERD SVLSTGSAAS HASTLSLASS QASGPTLSRQ LLTSFVSGLS DGVDSGYMED IEES AYERP RRPGGHERRG HRRPGQKFNR IYKLFKSTSQ MVLRRDSRSL EGSPDSGPPL RRAGSLCSPL DSPTLPPSRA QRSRS LPQP KLSPQLPGWL LAPASRHQRR RPFLSGDEDP KASTLRVVVF GSDRISGKVA RAYSNLRRLE NNRPLLTRFF KLQFFY VPV KRSRGTGTPT SPAPRSQTPP LPTDAPRHPG PAELGAAPWE ESTNDISHYL GMLDPWYERN VLGLMHLPPE VLCQSLK AE PRPLEGSPAQ LPILADMLLY YCRFAARPVL LQVYQTELTF ITGEKTTEIF IHSLELGHSA ATRAIKASGP GSKRLGID G DREAVPLTLQ IIYSKGAISG RSRWSNMEKL CTSVNLSKAC RQQEELDSST EALTLNLTEV VKRQTPKSKK GFNQISTSQ IKVDKVQIIG SNSCPFAVCL DQDERKILQS VIRCEVSPCY KPEKSSLCPP PQRPSYPPAP ATPDLCSLLC LPIMTFSGAL P UniProtKB: Phosphoinositide 3-kinase regulatory subunit 5 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.45 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8.5 Component:
Details: Freshly prepared gel filtration buffer, filtered through 0.22um filter and degassed | |||||||||||||||
| Grid | Model: C-flat-2/2 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa Details: Glow discharged using the Pelco EasiGlow. 15mA Current. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 1.5s blot time, -5 blot force. | |||||||||||||||
| Details | Specimen was a 1:1:1 molar ratio of p110g-p101-nanobody, purified to homogeneity by gel filtration. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 6808 / Average electron dose: 36.4 e/Å2 Details: Movies were collected in super-resolution mode set to collect 3 shots per grid hole over 9 holes by beam-shift before applying a stage shift. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)