+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22441 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The bacteriophage Phi-29 viral genome packaging motor assembly | |||||||||
 Map data Map data | Focused asymmetric reconstruction of the bacteriophage phi29 dsDNA packaging motor assembly at 4.1 A. | |||||||||
 Sample Sample | Bacillus virus phi29 viral genome packaging motor assembly != Bacillus virus phi29 Bacillus virus phi29 viral genome packaging motor assembly
| |||||||||
 Keywords Keywords | packaging motor / ATPase / MOTOR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral DNA genome packaging / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / ATP hydrolysis activity / DNA binding / RNA binding / ATP binding Similarity search - Function | |||||||||
| Biological species |   Bacillus phage phi29 (virus) / Bacillus phage phi29 (virus) /  Bacillus virus phi29 Bacillus virus phi29 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | White MA / Woodson M | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2021 Journal: Sci Adv / Year: 2021Title: A viral genome packaging motor transitions between cyclic and helical symmetry to translocate dsDNA. Authors: Michael Woodson / Joshua Pajak / Bryon P Mahler / Wei Zhao / Wei Zhang / Gaurav Arya / Mark A White / Paul J Jardine / Marc C Morais /  Abstract: Molecular segregation and biopolymer manipulation require the action of molecular motors to do work by applying directional forces to macromolecules. The additional strand conserved E (ASCE) ring ...Molecular segregation and biopolymer manipulation require the action of molecular motors to do work by applying directional forces to macromolecules. The additional strand conserved E (ASCE) ring motors are an ancient family of molecular motors responsible for diverse biological polymer manipulation tasks. Viruses use ASCE segregation motors to package their genomes into their protein capsids and provide accessible experimental systems due to their relative simplicity. We show by cryo-EM-focused image reconstruction that ASCE ATPases in viral double-stranded DNA (dsDNA) packaging motors adopt helical symmetry complementary to their dsDNA substrates. Together with previous data, our results suggest that these motors cycle between helical and planar configurations, providing a possible mechanism for directional translocation of DNA. Similar changes in quaternary structure have been observed for proteasome and helicase motors, suggesting an ancient and common mechanism of force generation that has been adapted for specific tasks over the course of evolution. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22441.map.gz emd_22441.map.gz | 113.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22441-v30.xml emd-22441-v30.xml emd-22441.xml emd-22441.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
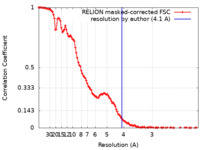
| FSC (resolution estimation) |  emd_22441_fsc.xml emd_22441_fsc.xml | 11.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_22441.png emd_22441.png | 251.7 KB | ||
| Filedesc metadata |  emd-22441.cif.gz emd-22441.cif.gz | 6.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22441 http://ftp.pdbj.org/pub/emdb/structures/EMD-22441 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22441 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22441 | HTTPS FTP |
-Related structure data
| Related structure data |  7jqqMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22441.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22441.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused asymmetric reconstruction of the bacteriophage phi29 dsDNA packaging motor assembly at 4.1 A. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Bacillus virus phi29 viral genome packaging motor assembly
| Entire | Name: Bacillus virus phi29 viral genome packaging motor assembly |
|---|---|
| Components |
|
-Supramolecule #1: Bacillus virus phi29
| Supramolecule | Name: Bacillus virus phi29 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 / NCBI-ID: 10756 / Sci species name: Bacillus virus phi29 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
-Macromolecule #1: DNA packaging protein
| Macromolecule | Name: DNA packaging protein / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:   Bacillus phage phi29 (virus) Bacillus phage phi29 (virus) |
| Molecular weight | Theoretical: 39.010406 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDKSLFYNPQ KMLSYDRILN FVIGARGIGK SYAMKVYPIN RFIKYGEQFI YVRRYKPELA KVSNYFNDVA QEFPDHELVV KGRRFYIDG KLAGWAIPLS VWQSEKSNAY PNVSTIVFDE FIREKDNSNY IPNEVSALLN LMDTVFRNRE RVRCICLSNA V SVVNPYFL ...String: MDKSLFYNPQ KMLSYDRILN FVIGARGIGK SYAMKVYPIN RFIKYGEQFI YVRRYKPELA KVSNYFNDVA QEFPDHELVV KGRRFYIDG KLAGWAIPLS VWQSEKSNAY PNVSTIVFDE FIREKDNSNY IPNEVSALLN LMDTVFRNRE RVRCICLSNA V SVVNPYFL FFNLVPDVNK RFNVYDDALI EIPDSLDFSS ERRKTRFGRL IDGTEYGEMS LDNQFIGDSQ VFIEKRSKDS KF VFSIVYN GFTLGVWVDV NQGLMYIDTA HDPSTKNVYT LTTDDLNENM MLITNYKNNY HLRKLASAFM NGYLRFDNQV IRN IAYELF RKMRIQ UniProtKB: DNA packaging protein |
-Macromolecule #2: pRNA (117-MER)
| Macromolecule | Name: pRNA (117-MER) / type: rna / ID: 2 / Number of copies: 5 |
|---|---|
| Source (natural) | Organism:  Bacillus virus phi29 Bacillus virus phi29 |
| Molecular weight | Theoretical: 37.469012 KDa |
| Sequence | String: GGAAUGGUAC GGUACUUCCA UUGUCAUGUG UAUGUUGGGG AUUAAACCCU GAUUGAGUUC AGCCCACAUA CUUUGUUGAU UGGUUGUCA AUCAUGGCAA AAGUGCACGC UACUUUCC |
-Macromolecule #3: DNA (60-MER)
| Macromolecule | Name: DNA (60-MER) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Bacillus virus phi29 Bacillus virus phi29 |
| Molecular weight | Theoretical: 18.491848 KDa |
| Sequence | String: (DG)(DT)(DC)(DA)(DG)(DT)(DC)(DA)(DG)(DT) (DC)(DA)(DG)(DT)(DC)(DA)(DG)(DT)(DC)(DA) (DG)(DT)(DC)(DA)(DG)(DT)(DC)(DA)(DG) (DT)(DC)(DA)(DG)(DT)(DC)(DA)(DG)(DT)(DC) (DA) (DG)(DT)(DC)(DA)(DG)(DT) ...String: (DG)(DT)(DC)(DA)(DG)(DT)(DC)(DA)(DG)(DT) (DC)(DA)(DG)(DT)(DC)(DA)(DG)(DT)(DC)(DA) (DG)(DT)(DC)(DA)(DG)(DT)(DC)(DA)(DG) (DT)(DC)(DA)(DG)(DT)(DC)(DA)(DG)(DT)(DC) (DA) (DG)(DT)(DC)(DA)(DG)(DT)(DC)(DA) (DG)(DT)(DC)(DA)(DG)(DT)(DC)(DA)(DG)(DT) (DC)(DA) |
-Macromolecule #4: DNA (60-MER)
| Macromolecule | Name: DNA (60-MER) / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Bacillus virus phi29 Bacillus virus phi29 |
| Molecular weight | Theoretical: 18.49185 KDa |
| Sequence | String: (DT)(DG)(DA)(DC)(DT)(DG)(DA)(DC)(DT)(DG) (DA)(DC)(DT)(DG)(DA)(DC)(DT)(DG)(DA)(DC) (DT)(DG)(DA)(DC)(DT)(DG)(DA)(DC)(DT) (DG)(DA)(DC)(DT)(DG)(DA)(DC)(DT)(DG)(DA) (DC) (DT)(DG)(DA)(DC)(DT)(DG) ...String: (DT)(DG)(DA)(DC)(DT)(DG)(DA)(DC)(DT)(DG) (DA)(DC)(DT)(DG)(DA)(DC)(DT)(DG)(DA)(DC) (DT)(DG)(DA)(DC)(DT)(DG)(DA)(DC)(DT) (DG)(DA)(DC)(DT)(DG)(DA)(DC)(DT)(DG)(DA) (DC) (DT)(DG)(DA)(DC)(DT)(DG)(DA)(DC) (DT)(DG)(DA)(DC)(DT)(DG)(DA)(DC)(DT)(DG) (DA)(DC) |
-Macromolecule #5: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / type: ligand / ID: 5 / Number of copies: 3 / Formula: AGS |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 3 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF 2002 |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 7676 pixel / Digitization - Dimensions - Height: 7420 pixel / Digitization - Frames/image: 0-44 / Average electron dose: 41.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 4.2 µm / Calibrated defocus min: 1.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.2 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)