[English] 日本語
 Yorodumi
Yorodumi- EMDB-2226: Single particle analysis of recombinant human anaphase promoting ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2226 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single particle analysis of recombinant human anaphase promoting complex (APC/C) | |||||||||
 Map data Map data | reconstruction of recombinant human APC/C | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Anaphase promoting complex / cyclosome / cell cycle / recombinant expression / single particle / 3-dimensional structure / ubiquitination | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 20.0 Å | |||||||||
 Authors Authors | Zhang Z / Yang J / Kong EH / Chao WCH / Morris EP / da Fonseca PCA / Barford D | |||||||||
 Citation Citation |  Journal: Biochem J / Year: 2013 Journal: Biochem J / Year: 2013Title: Recombinant expression, reconstitution and structure of human anaphase-promoting complex (APC/C). Authors: Ziguo Zhang / Jing Yang / Eric H Kong / William C H Chao / Edward P Morris / Paula C A da Fonseca / David Barford /  Abstract: Mechanistic and structural studies of large multi-subunit assemblies are greatly facilitated by their reconstitution in heterologous recombinant systems. In the present paper, we describe the ...Mechanistic and structural studies of large multi-subunit assemblies are greatly facilitated by their reconstitution in heterologous recombinant systems. In the present paper, we describe the generation of recombinant human APC/C (anaphase-promoting complex/cyclosome), an E3 ubiquitin ligase that regulates cell-cycle progression. Human APC/C is composed of 14 distinct proteins that assemble into a complex of at least 19 subunits with a combined molecular mass of ~1.2 MDa. We show that recombinant human APC/C is correctly assembled, as judged by its capacity to ubiquitinate the budding yeast APC/C substrate Hsl1 (histone synthetic lethal 1) dependent on the APC/C co-activator Cdh1 [Cdc (cell division cycle) 20 homologue 1], and its three-dimensional reconstruction by electron microscopy and single-particle analysis. Successful reconstitution validates the subunit composition of human APC/C. The structure of human APC/C is compatible with the Saccharomyces cerevisiae APC/C homology model, and in contrast with endogenous human APC/C, no evidence for conformational flexibility of the TPR (tetratricopeptide repeat) lobe is observed. Additional density present in the human APC/C structure, proximal to Apc3/Cdc27 of the TPR lobe, is assigned to the TPR subunit Apc7, a subunit specific to vertebrate APC/C. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2226.map.gz emd_2226.map.gz | 10 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2226-v30.xml emd-2226-v30.xml emd-2226.xml emd-2226.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2226.tif emd_2226.tif | 137.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2226 http://ftp.pdbj.org/pub/emdb/structures/EMD-2226 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2226 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2226 | HTTPS FTP |
-Validation report
| Summary document |  emd_2226_validation.pdf.gz emd_2226_validation.pdf.gz | 197.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2226_full_validation.pdf.gz emd_2226_full_validation.pdf.gz | 196.2 KB | Display | |
| Data in XML |  emd_2226_validation.xml.gz emd_2226_validation.xml.gz | 5.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2226 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2226 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2226 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2226 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2226.map.gz / Format: CCP4 / Size: 10.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2226.map.gz / Format: CCP4 / Size: 10.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | reconstruction of recombinant human APC/C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.4725 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
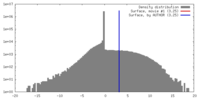
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : recombinant human APC/C
+Supramolecule #1000: recombinant human APC/C
+Macromolecule #1: Apc1
+Macromolecule #2: Apc5
+Macromolecule #3: Apc15
+Macromolecule #4: Apc10
+Macromolecule #5: Apc8
+Macromolecule #6: Apc13
+Macromolecule #7: Apc2
+Macromolecule #8: Apc4
+Macromolecule #9: Apc11
+Macromolecule #10: Apc3
+Macromolecule #11: Apc16
+Macromolecule #12: Cdc26
+Macromolecule #13: Apc7
+Macromolecule #14: Apc6
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 / Details: 20 mM HEPES NaOH pH 8.0, 200 mM NaCl, 2 mM DTT |
|---|---|
| Staining | Type: NEGATIVE / Details: grids stained with 2% (w/v) uranyl acetate |
| Grid | Details: Quantifoil R1.2/1.3 grids coated with a thin layer of carbon |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Date | Jan 1, 2012 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F415 (4k x 4k) / Average electron dose: 100 e/Å2 Details: on recording each adjacent boxes of 2x2 pixels were averaged |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus min: 1.2 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Particles were manually selected using the EMAN boxer software. The images were analysed using a combination of IMAGIC and Spider software. |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Resolution method: OTHER / Software - Name: Spider, Imagic / Number images used: 5639 |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)



















 Trichoplusia ni (cabbage looper)
Trichoplusia ni (cabbage looper)