[English] 日本語
 Yorodumi
Yorodumi- EMDB-20857: CryoEM structure of human alpha4beta2 nicotinic acetylcholine rec... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20857 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor in complex with varenicline | |||||||||
 Map data Map data | cytochrome B562-fused human alpha4beta2 nicotinic acetylcholine receptor in complex with fab fragments and bound to varenicline | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationvestibulocochlear nerve development / lateral geniculate nucleus development / regulation of circadian sleep/wake cycle, REM sleep / regulation of synaptic transmission, dopaminergic / synaptic transmission involved in micturition / quaternary ammonium group binding / Highly sodium permeable postsynaptic acetylcholine nicotinic receptors / Highly calcium permeable nicotinic acetylcholine receptors / optic nerve morphogenesis / central nervous system projection neuron axonogenesis ...vestibulocochlear nerve development / lateral geniculate nucleus development / regulation of circadian sleep/wake cycle, REM sleep / regulation of synaptic transmission, dopaminergic / synaptic transmission involved in micturition / quaternary ammonium group binding / Highly sodium permeable postsynaptic acetylcholine nicotinic receptors / Highly calcium permeable nicotinic acetylcholine receptors / optic nerve morphogenesis / central nervous system projection neuron axonogenesis / Highly calcium permeable postsynaptic nicotinic acetylcholine receptors / acetylcholine receptor activity / response to acetylcholine / cholinergic synapse / acetylcholine-gated channel complex / negative regulation of action potential / positive regulation of dopamine secretion / regulation of dopamine metabolic process / behavioral response to nicotine / acetylcholine-gated monoatomic cation-selective channel activity / inhibitory postsynaptic potential / nervous system process / synaptic transmission, cholinergic / acetylcholine binding / acetylcholine receptor signaling pathway / postsynaptic specialization membrane / heterocyclic compound binding / regulation of synapse assembly / regulation of dendrite morphogenesis / action potential / regulation of dopamine secretion / plasma membrane raft / B cell activation / associative learning / membrane depolarization / social behavior / ligand-gated monoatomic ion channel activity / smooth muscle contraction / monoatomic ion transport / positive regulation of B cell proliferation / sensory perception of pain / visual perception / regulation of membrane potential / learning / response to cocaine / locomotory behavior / response to nicotine / sensory perception of sound / electron transport chain / visual learning / memory / cognition / calcium ion transport / presynaptic membrane / chemical synaptic transmission / postsynaptic membrane / response to ethanol / response to oxidative stress / periplasmic space / electron transfer activity / response to hypoxia / neuron projection / iron ion binding / external side of plasma membrane / DNA repair / neuronal cell body / dendrite / heme binding / synapse / protein-containing complex binding / signal transduction / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
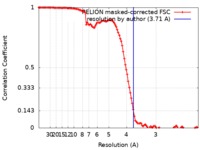
| Method | single particle reconstruction / cryo EM / Resolution: 3.71 Å | |||||||||
 Authors Authors | Alvarez FJD / Mukherjee S / Han S / Ammirati M / Kossiakoff AA | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins. Authors: Somnath Mukherjee / Satchal K Erramilli / Mark Ammirati / Frances J D Alvarez / Kimberly F Fennell / Michael D Purdy / Blazej M Skrobek / Katarzyna Radziwon / John Coukos / Yanyong Kang / ...Authors: Somnath Mukherjee / Satchal K Erramilli / Mark Ammirati / Frances J D Alvarez / Kimberly F Fennell / Michael D Purdy / Blazej M Skrobek / Katarzyna Radziwon / John Coukos / Yanyong Kang / Przemysław Dutka / Xiang Gao / Xiayang Qiu / Mark Yeager / H Eric Xu / Seungil Han / Anthony A Kossiakoff /    Abstract: We propose the concept of universal fiducials based on a set of pre-made semi-synthetic antibodies (sABs) generated by customized phage display selections against the fusion protein BRIL, an ...We propose the concept of universal fiducials based on a set of pre-made semi-synthetic antibodies (sABs) generated by customized phage display selections against the fusion protein BRIL, an engineered variant of apocytochrome b562a. These sABs can bind to BRIL fused either into the loops or termini of different GPCRs, ion channels, receptors and transporters without disrupting their structure. A crystal structure of BRIL in complex with an affinity-matured sAB (BAG2) that bound to all systems tested delineates the footprint of interaction. Negative stain and cryoEM data of several examples of BRIL-membrane protein chimera highlight the effectiveness of the sABs as universal fiducial marks. Taken together with a cryoEM structure of sAB bound human nicotinic acetylcholine receptor, this work demonstrates that these anti-BRIL sABs can greatly enhance the particle properties leading to improved cryoEM outcomes, especially for challenging membrane proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20857.map.gz emd_20857.map.gz | 8.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20857-v30.xml emd-20857-v30.xml emd-20857.xml emd-20857.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20857_fsc.xml emd_20857_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_20857.png emd_20857.png | 139.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20857 http://ftp.pdbj.org/pub/emdb/structures/EMD-20857 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20857 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20857 | HTTPS FTP |
-Validation report
| Summary document |  emd_20857_validation.pdf.gz emd_20857_validation.pdf.gz | 344.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20857_full_validation.pdf.gz emd_20857_full_validation.pdf.gz | 344.1 KB | Display | |
| Data in XML |  emd_20857_validation.xml.gz emd_20857_validation.xml.gz | 12.2 KB | Display | |
| Data in CIF |  emd_20857_validation.cif.gz emd_20857_validation.cif.gz | 16.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20857 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20857 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20857 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20857 | HTTPS FTP |
-Related structure data
| Related structure data |  6ur8MC  6cbvC  6usfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20857.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20857.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cytochrome B562-fused human alpha4beta2 nicotinic acetylcholine receptor in complex with fab fragments and bound to varenicline | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1024 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : human alpha4beta2 nicotinic acetylcholine receptor in complex fab...
| Entire | Name: human alpha4beta2 nicotinic acetylcholine receptor in complex fab fragments and bound to varenicline |
|---|---|
| Components |
|
-Supramolecule #1: human alpha4beta2 nicotinic acetylcholine receptor in complex fab...
| Supramolecule | Name: human alpha4beta2 nicotinic acetylcholine receptor in complex fab fragments and bound to varenicline type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Chimera of soluble cytochrome b562 (BRIL) and neuronal acetylchol...
| Macromolecule | Name: Chimera of soluble cytochrome b562 (BRIL) and neuronal acetylcholine receptor subunit alpha-4 type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 56.027848 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SSHVETRAHA EERLLKKLFS GYNKWSRPVA NISDVVLVRF GLSIAQLIDV DEKNQMMTTN VWVKQEWHDY KLRWDPADYE NVTSIRIPS ELIWRPDIVL YNNADGDFAV THLTKAHLFH DGRVQWTPPA IYKSSCSIDV TFFPFDQQNC TMKFGSWTYD K AKIDLVNM ...String: SSHVETRAHA EERLLKKLFS GYNKWSRPVA NISDVVLVRF GLSIAQLIDV DEKNQMMTTN VWVKQEWHDY KLRWDPADYE NVTSIRIPS ELIWRPDIVL YNNADGDFAV THLTKAHLFH DGRVQWTPPA IYKSSCSIDV TFFPFDQQNC TMKFGSWTYD K AKIDLVNM HSRVDQLDFW ESGEWVIVDA VGTYNTRKYE CCAEIYPDIT YAFVIRRLPL FYTINLIIPC LLISCLTVLV FY LPSECGE KITLCISVLL SLTVFLLLIT EIIPSTSLVI PLIGEYLLFT MIFVTLSIVI TVFVLNVHHR SPRTHTMPTW VRR VFLDIV PRLLLMKRPS VVDTDFADLE DNWETLNDNL KVIEKADNAA QVKDALTKMR AAALDAQKAT PPKLEDKSPD SPEM KDFRH GFDILVGQID DALKLANEGK VKEAQAAAEQ LKTTRNAYIQ KYLEDWKYVA MVIDRIFLWM FIIVCLLGTV GLFLP PWLA GMI |
-Macromolecule #2: Neuronal acetylcholine receptor subunit beta-2
| Macromolecule | Name: Neuronal acetylcholine receptor subunit beta-2 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.931953 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: TDTEERLVEH LLDPSRYNKL IRPATNGSEL VTVQLMVSLA QLISVHEREQ IMTTNVWLTQ EWEDYRLTWK PEEFDNMKKV RLPSKHIWL PDVVLYNNAD GMYEVSFYSN AVVSYDGSIF WLPPAIYKSA CKIEVKHFPF DQQNCTMKFR SWTYDRTEID L VLKSEVAS ...String: TDTEERLVEH LLDPSRYNKL IRPATNGSEL VTVQLMVSLA QLISVHEREQ IMTTNVWLTQ EWEDYRLTWK PEEFDNMKKV RLPSKHIWL PDVVLYNNAD GMYEVSFYSN AVVSYDGSIF WLPPAIYKSA CKIEVKHFPF DQQNCTMKFR SWTYDRTEID L VLKSEVAS LDDFTPSGEW DIVALPGRRN ENPDDSTYVD ITYDFIIRRK PLFYTINLII PCVLITSLAI LVFYLPSDCG EK MTLCISV LLALTVFLLL ISKIVPPTSL DVPLVGKYLM FTMVLVTFSI VTSVCVLNVH HRSPTTHTMA PWVKVVFLEK LPA LLFMQQ DDDQSVSEDW KYVAMVIDRL FLWIFVFVCV FGTIGMFLQP LFQNYTTTTF LHSDHSAPSS KSAWSHPQFE K |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 8 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: VARENICLINE
| Macromolecule | Name: VARENICLINE / type: ligand / ID: 4 / Number of copies: 2 / Formula: QMR |
|---|---|
| Molecular weight | Theoretical: 211.262 Da |
| Chemical component information | 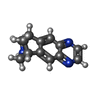 ChemComp-QMR: |
-Macromolecule #5: beta-D-mannopyranose
| Macromolecule | Name: beta-D-mannopyranose / type: ligand / ID: 5 / Number of copies: 3 / Formula: BMA |
|---|---|
| Molecular weight | Theoretical: 180.156 Da |
| Chemical component information |  ChemComp-BMA: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.019 kPa | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Average exposure time: 10.5 sec. / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)