[English] 日本語
 Yorodumi
Yorodumi- EMDB-20398: In situ structure of BTV RNA-dependent RNA polymerase in BTV virion -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20398 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ structure of BTV RNA-dependent RNA polymerase in BTV virion | ||||||||||||||||||||||||
 Map data Map data | BTV RNA-dependent RNA polymerase in BTV virion | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | RNA dependent RNA polymerase / Viral protein / Transferase | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationviral genome replication / RNA-directed RNA polymerase / RNA-dependent RNA polymerase activity / nucleotide binding / DNA-templated transcription / structural molecule activity / RNA binding Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  Bluetongue virus 1 Bluetongue virus 1 | ||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | ||||||||||||||||||||||||
 Authors Authors | He Y / Shivakoti S | ||||||||||||||||||||||||
| Funding support |  United States, United States,  United Kingdom, 7 items United Kingdom, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2019 Journal: Proc Natl Acad Sci U S A / Year: 2019Title: In situ structures of RNA-dependent RNA polymerase inside bluetongue virus before and after uncoating. Authors: Yao He / Sakar Shivakoti / Ke Ding / Yanxiang Cui / Polly Roy / Z Hong Zhou /   Abstract: Bluetongue virus (BTV), a major threat to livestock, is a multilayered, nonturreted member of the , a family of segmented dsRNA viruses characterized by endogenous RNA transcription through an RNA- ...Bluetongue virus (BTV), a major threat to livestock, is a multilayered, nonturreted member of the , a family of segmented dsRNA viruses characterized by endogenous RNA transcription through an RNA-dependent RNA polymerase (RdRp). To date, the structure of BTV RdRp has been unknown, limiting our mechanistic understanding of BTV transcription and hindering rational drug design effort targeting this essential enzyme. Here, we report the in situ structures of BTV RdRp VP1 in both the triple-layered virion and double-layered core, as determined by cryo-electron microscopy (cryoEM) and subparticle reconstruction. BTV RdRp has 2 unique motifs not found in other viral RdRps: a fingernail, attached to the conserved fingers subdomain, and a bundle of 3 helices: 1 from the palm subdomain and 2 from the N-terminal domain. BTV RdRp VP1 is anchored to the inner surface of the capsid shell via 5 asymmetrically arranged N termini of the inner capsid shell protein VP3A around the 5-fold axis. The structural changes of RdRp VP1 and associated capsid shell proteins between BTV virions and cores suggest that the detachment of the outer capsid proteins VP2 and VP5 during viral entry induces both global movements of the inner capsid shell and local conformational changes of the N-terminal latch helix (residues 34 to 51) of 1 inner capsid shell protein VP3A, priming RdRp VP1 within the capsid for transcription. Understanding this mechanism in BTV also provides general insights into RdRp activation and regulation during viral entry of other multilayered, nonturreted dsRNA viruses. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20398.map.gz emd_20398.map.gz | 87 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20398-v30.xml emd-20398-v30.xml emd-20398.xml emd-20398.xml | 13.6 KB 13.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20398.png emd_20398.png | 227.8 KB | ||
| Filedesc metadata |  emd-20398.cif.gz emd-20398.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20398 http://ftp.pdbj.org/pub/emdb/structures/EMD-20398 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20398 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20398 | HTTPS FTP |
-Validation report
| Summary document |  emd_20398_validation.pdf.gz emd_20398_validation.pdf.gz | 704.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20398_full_validation.pdf.gz emd_20398_full_validation.pdf.gz | 704.3 KB | Display | |
| Data in XML |  emd_20398_validation.xml.gz emd_20398_validation.xml.gz | 6.5 KB | Display | |
| Data in CIF |  emd_20398_validation.cif.gz emd_20398_validation.cif.gz | 7.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20398 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20398 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20398 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20398 | HTTPS FTP |
-Related structure data
| Related structure data |  6pnsMC  6po2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20398.map.gz / Format: CCP4 / Size: 96.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20398.map.gz / Format: CCP4 / Size: 96.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BTV RNA-dependent RNA polymerase in BTV virion | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
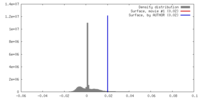
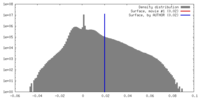
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Bluetongue virus 1
| Entire | Name:  Bluetongue virus 1 Bluetongue virus 1 |
|---|---|
| Components |
|
-Supramolecule #1: Bluetongue virus 1
| Supramolecule | Name: Bluetongue virus 1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 35327 / Sci species name: Bluetongue virus 1 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: RNA-directed RNA polymerase
| Macromolecule | Name: RNA-directed RNA polymerase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Bluetongue virus 1 Bluetongue virus 1 |
| Molecular weight | Theoretical: 149.926281 KDa |
| Sequence | String: MVAITVQGAQ LIKRVVERFY PGIAFNINEG ACYIYKFSDH IRRIRMKHGT KYRRQAEEII RNISLRKERL YGIPVLDEVE WKYVFDGQT FQSYAFEVYV NSILPWSELD PEEEFLRNYR VSREMTEVEK FIEFRAKNEM QIYGDIPIKV WCCFINELSA E LKHVPLGM ...String: MVAITVQGAQ LIKRVVERFY PGIAFNINEG ACYIYKFSDH IRRIRMKHGT KYRRQAEEII RNISLRKERL YGIPVLDEVE WKYVFDGQT FQSYAFEVYV NSILPWSELD PEEEFLRNYR VSREMTEVEK FIEFRAKNEM QIYGDIPIKV WCCFINELSA E LKHVPLGM QVMADFVNRF DSPFHQGNRD LSNLEDFQVA YTTPLLFEMC CMESILEFNI KMRMREEEIS ALEFGDMKVD PV GLLREFF ILCLPHPKKI NNVLRAPYSW FVKMWGVGAD PIVVLQSTAG DDRNSKDVFY DKFRTEPNRY KALFRSSFYN ESR RMNEEK ILEAVKYSQK LGSHDRRLPL FEKMLKTVYT TPFYPHKSSN MILASFLLSI QTITGYGRAW VKNVSTEFDK QLKP NPSNL VQDVSDLTRE FFKQAYVEAK ERREEIVKPE DLYTSMLRLA RNTSSGFSTE IYVKKRFGPR LRDKDLIKIN SRIKA LVIF TKGHTVFTDE ELHKKYNSVE LYQTKGSRDV PIKATRTIYS INLSVLVPQL IVTLPLNEYF SRVGGITSPD YKKIGG KVI VGDLEATGSR VMDAADCFRN SADRDIFTIA IDYSEYDTHL TRHNFRTGML QGIREAMAPY RDLRYEGYTL EQIIDFG YG EGRVANTLWN GKRRLFKTTF DAYIRLDESE RDKGSFKVPK GVLPVSSVDV ANRIAVDKGF DTLIAATDGS DLALIDTH L SGENSTLIAN SMHNMAIGTL MQREVGREQP GVLTFLSEQY VGDDTLFYTK LHTTDTKVFD KVAASIFDTV AKCGHEASP SKTMMTPYSV EKTQTHAKQG CYVPQDRMMI ISSERRKDIE DVQGYVRSQV QTMITKVSRG FCHDLAQLIL MLKTTFIGAW KMKRTIKED AMYRDRKFDS NDEDGFTLIQ IRNPLALYVP IGWNGYGAHP AALNIVMTEE MYVDSIMISK LDEIMAPIRR I VHDIPPCW NETQGDKRGL ISATKMSFFS KMARPAVQAA LSDPQIINLV EELPLGEFSP GRISRTMMHS ALLKESSART LL SSGYELE YQKALNSWIT QVSMRLGEES GVISTSYAKL FDVYFEGELD GAPHMFPDQN LSPQFYIQKM MIGPRVSSRV RNS YVDRID VILRKDVVMR GFITANTILN VIEKLGTNHS VGDLVTVFTL MNIETRVAEE LAEYMTSEKI RFDALKLLKK GIAG DEFTM SLNVATQDFI DTYLAYPYQL TKTEVDAISL YCTQMIMLRA ALGLPKKKMK IVVTDDAKKR YKIRLQRFRT HVPKI KVLK KLIDPNRMTV RNLENQFV UniProtKB: RNA-directed RNA polymerase |
-Macromolecule #2: Inner core structural protein VP3
| Macromolecule | Name: Inner core structural protein VP3 / type: protein_or_peptide / ID: 2 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bluetongue virus 1 Bluetongue virus 1 |
| Molecular weight | Theoretical: 103.410508 KDa |
| Sequence | String: MAAQNEQRPE RIKTTPYLEG DVLSSDSGPL LSVFALQEIM QKVRQVQADY MTATREVDFT VPDVQKILDD IKALAAEQVY KIVKVPSIS FRHIVMQSRD RVLRVDTYYE EMSQVGDVIT EDEPEKFYST IIKKVRFIRG KGSFILHDIP TRDHRGMEVA E PEVLGVEF ...String: MAAQNEQRPE RIKTTPYLEG DVLSSDSGPL LSVFALQEIM QKVRQVQADY MTATREVDFT VPDVQKILDD IKALAAEQVY KIVKVPSIS FRHIVMQSRD RVLRVDTYYE EMSQVGDVIT EDEPEKFYST IIKKVRFIRG KGSFILHDIP TRDHRGMEVA E PEVLGVEF KNVLPVLTAE HRAMIQNALD GSIIENGNVA TRDVDVFIGA CSEPVYRIYN RLQGYIEAVQ LQELRNSIGW LE RLGHRKR ITYSQEVLTD FRRQDTIWVL ALQLPVNPQV VWDVPRSSIA NLIMNIATCL PTGEYIAPNP RISSITLTQR ITT TGPFAI LTGSTPTAQQ LNDVRKIYLA LMFPGQIILD LKIDPGERMD PAVRMVAGVV GHLLFTAGGR FTNLTQNMAR QLDI ALNDY LLYMYNTRVQ VNYGPTGEPL DFQIGRNQYD CNVFRADFAT GTGYNGWATI DVEYREPAPY VHAQRYIRYC GIDSR ELIN PTTYGIGMTY HCYNEMLRML VAAGKDSEAA YFRSMLPFHM VRFARINQII NEDLHSVFSL PDDMFNALLP DLIAGA HQN ADPVVLDVSW ISLWFAFNRS FEPTHRNEML EVAPLIESVY ASELSVMKVD MRHLSLMQRR FPDVLIQARP SHFWKAV LN DSPEAVKAVM NLSHSHNFIN IRDMMRWVML PSLQPSLKLA LEEEAWAAAN DFEDLMLTDQ VYMHRDMLPE PRLDDIER F RQEGFYYTNM LEAPPEIDRV VQYTYEIARL QANMGQFRAA LRRIMDDDDW VRFGGVLRTV RVKFYDARPP DDVLQGLPF SYDTNERGGL AYATIKYATE TTIFYLIYNV EFSNTPDSLV LINPTYTMTK VFINKRIVER VRVGQILAVL NRRFVAYKGK MRIMDITQS LKMGTKLAAP TV UniProtKB: Core protein VP3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8.8 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 32.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.7 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 244063 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)