[English] 日本語
 Yorodumi
Yorodumi- EMDB-20378: CryoEM structure of zebra fish alpha-1 glycine receptor bound wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20378 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of zebra fish alpha-1 glycine receptor bound with GABA in SMA, super-open state | |||||||||
 Map data Map data | unsharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | glycine receptor / SMA / CryoEM / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationNeurotransmitter receptors and postsynaptic signal transmission / extracellularly glycine-gated ion channel activity / extracellularly glycine-gated chloride channel activity / transmitter-gated monoatomic ion channel activity / regulation of neuron differentiation / glycine binding / cellular response to zinc ion / ligand-gated monoatomic ion channel activity / cellular response to ethanol / response to amino acid ...Neurotransmitter receptors and postsynaptic signal transmission / extracellularly glycine-gated ion channel activity / extracellularly glycine-gated chloride channel activity / transmitter-gated monoatomic ion channel activity / regulation of neuron differentiation / glycine binding / cellular response to zinc ion / ligand-gated monoatomic ion channel activity / cellular response to ethanol / response to amino acid / chloride channel complex / monoatomic ion transport / chloride transmembrane transport / central nervous system development / cellular response to amino acid stimulus / transmembrane signaling receptor activity / perikaryon / postsynaptic membrane / dendrite / zinc ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Yu J / Zhu H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2021 Journal: Cell / Year: 2021Title: Mechanism of gating and partial agonist action in the glycine receptor. Authors: Jie Yu / Hongtao Zhu / Remigijus Lape / Timo Greiner / Juan Du / Wei Lü / Lucia Sivilotti / Eric Gouaux /   Abstract: Ligand-gated ion channels mediate signal transduction at chemical synapses and transition between resting, open, and desensitized states in response to neurotransmitter binding. Neurotransmitters ...Ligand-gated ion channels mediate signal transduction at chemical synapses and transition between resting, open, and desensitized states in response to neurotransmitter binding. Neurotransmitters that produce maximum open channel probabilities (Po) are full agonists, whereas those that yield lower than maximum Po are partial agonists. Cys-loop receptors are an important class of neurotransmitter receptors, yet a structure-based understanding of the mechanism of partial agonist action has proven elusive. Here, we study the glycine receptor with the full agonist glycine and the partial agonists taurine and γ-amino butyric acid (GABA). We use electrophysiology to show how partial agonists populate agonist-bound, closed channel states and cryo-EM reconstructions to illuminate the structures of intermediate, pre-open states, providing insights into previously unseen conformational states along the receptor reaction pathway. We further correlate agonist-induced conformational changes to Po across members of the receptor family, providing a hypothetical mechanism for partial and full agonist action at Cys-loop receptors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20378.map.gz emd_20378.map.gz | 110.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20378-v30.xml emd-20378-v30.xml emd-20378.xml emd-20378.xml | 9.8 KB 9.8 KB | Display Display |  EMDB header EMDB header |
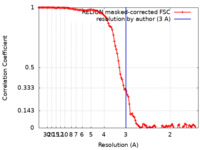
| FSC (resolution estimation) |  emd_20378_fsc.xml emd_20378_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_20378.png emd_20378.png | 76.1 KB | ||
| Filedesc metadata |  emd-20378.cif.gz emd-20378.cif.gz | 5.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20378 http://ftp.pdbj.org/pub/emdb/structures/EMD-20378 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20378 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20378 | HTTPS FTP |
-Related structure data
| Related structure data |  6plwMC  6ploC  6plpC  6plqC  6plrC  6plsC  6pltC  6pluC  6plvC  6plxC  6plyC  6plzC  6pm0C  6pm1C  6pm2C  6pm3C  6pm4C  6pm5C  6pm6C  6pxdC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20378.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20378.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.823 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
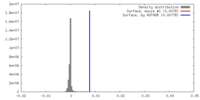
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : glycine receptor in complex with GABA
| Entire | Name: glycine receptor in complex with GABA |
|---|---|
| Components |
|
-Supramolecule #1: glycine receptor in complex with GABA
| Supramolecule | Name: glycine receptor in complex with GABA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 250 kDa/nm |
-Macromolecule #1: Glycine receptor subunit alphaZ1
| Macromolecule | Name: Glycine receptor subunit alphaZ1 / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 52.537598 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFALGIYLWE TIVFFSLAAS QQAAARKAAS PMPPSEFLDK LMGKVSGYDA RIRPNFKGPP VNVTCNIFIN SFGSIAETTM DYRVNIFLR QQWNDPRLAY SEYPDDSLDL DPSMLDSIWK PDLFFANEKG ANFHEVTTDN KLLRISKNGN VLYSIRITLV L ACPMDLKN ...String: MFALGIYLWE TIVFFSLAAS QQAAARKAAS PMPPSEFLDK LMGKVSGYDA RIRPNFKGPP VNVTCNIFIN SFGSIAETTM DYRVNIFLR QQWNDPRLAY SEYPDDSLDL DPSMLDSIWK PDLFFANEKG ANFHEVTTDN KLLRISKNGN VLYSIRITLV L ACPMDLKN FPMDVQTCIM QLESFGYTMN DLIFEWDEKG AVQVADGLTL PQFILKEEKD LRYCTKHYNT GKFTCIEARF HL ERQMGYY LIQMYIPSLL IVILSWVSFW INMDAAPARV GLGITTVLTM TTQSSGSRAS LPKVSYVKAI DIWMAVCLLF VFS ALLEYA AVNFIARQHK ELLRFQRRRR HLKEDEAGDG RFSFAAYGMG PACLQAKDGM AIKGNNNNAP TSTNPPEKTV EEMR KLFIS RAKRIDTVSR VAFPLVFLIF NIFYWITYKI IRSEDIHKQL VPRGSHHHHH HHH UniProtKB: Glycine receptor subunit alphaZ1 |
-Macromolecule #3: GAMMA-AMINO-BUTANOIC ACID
| Macromolecule | Name: GAMMA-AMINO-BUTANOIC ACID / type: ligand / ID: 3 / Number of copies: 5 / Formula: ABU |
|---|---|
| Molecular weight | Theoretical: 103.12 Da |
| Chemical component information |  ChemComp-ABU: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 40.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)