[English] 日本語
 Yorodumi
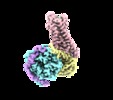
Yorodumi- EMDB-20181: human Neurotensin Receptor 1 (hNTSR1) - Gi1 Protein Complex in no... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20181 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | human Neurotensin Receptor 1 (hNTSR1) - Gi1 Protein Complex in non-canonical conformation (NC state) | |||||||||||||||||||||
 Map data Map data | human neurotensin receptor 1 (hNTSR1) - Gi1 complex in non-canonical conformation (NC state) | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | GPCR / G-protein / complex / SIGNALING PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationG protein-coupled neurotensin receptor activity / inositol phosphate catabolic process / symmetric synapse / D-aspartate import across plasma membrane / positive regulation of gamma-aminobutyric acid secretion / regulation of membrane depolarization / positive regulation of arachidonate secretion / L-glutamate import across plasma membrane / regulation of respiratory gaseous exchange / positive regulation of inhibitory postsynaptic potential ...G protein-coupled neurotensin receptor activity / inositol phosphate catabolic process / symmetric synapse / D-aspartate import across plasma membrane / positive regulation of gamma-aminobutyric acid secretion / regulation of membrane depolarization / positive regulation of arachidonate secretion / L-glutamate import across plasma membrane / regulation of respiratory gaseous exchange / positive regulation of inhibitory postsynaptic potential / negative regulation of systemic arterial blood pressure / negative regulation of release of sequestered calcium ion into cytosol / positive regulation of glutamate secretion / temperature homeostasis / response to lipid / positive regulation of inositol phosphate biosynthetic process / detection of temperature stimulus involved in sensory perception of pain / neuropeptide signaling pathway / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / D2 dopamine receptor binding / response to prostaglandin E / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cellular response to forskolin / Peptide ligand-binding receptors / positive regulation of release of sequestered calcium ion into cytosol / regulation of mitotic spindle organization / dendritic shaft / adult locomotory behavior / learning / Regulation of insulin secretion / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / response to peptide hormone / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / cytoplasmic side of plasma membrane / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / terminal bouton / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / GDP binding / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / G protein activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / midbody / cell cortex / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / perikaryon / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / dendritic spine / chemical synaptic transmission Similarity search - Function | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||||||||||||||
 Authors Authors | Kato HE / Zhang Y | |||||||||||||||||||||
| Funding support |  Japan, Japan,  Denmark, Denmark,  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2019 Journal: Nature / Year: 2019Title: Conformational transitions of a neurotensin receptor 1-G complex. Authors: Hideaki E Kato / Yan Zhang / Hongli Hu / Carl-Mikael Suomivuori / Francois Marie Ngako Kadji / Junken Aoki / Kaavya Krishna Kumar / Rasmus Fonseca / Daniel Hilger / Weijiao Huang / Naomi R ...Authors: Hideaki E Kato / Yan Zhang / Hongli Hu / Carl-Mikael Suomivuori / Francois Marie Ngako Kadji / Junken Aoki / Kaavya Krishna Kumar / Rasmus Fonseca / Daniel Hilger / Weijiao Huang / Naomi R Latorraca / Asuka Inoue / Ron O Dror / Brian K Kobilka / Georgios Skiniotis /     Abstract: Neurotensin receptor 1 (NTSR1) is a G-protein-coupled receptor (GPCR) that engages multiple subtypes of G protein, and is involved in the regulation of blood pressure, body temperature, weight and ...Neurotensin receptor 1 (NTSR1) is a G-protein-coupled receptor (GPCR) that engages multiple subtypes of G protein, and is involved in the regulation of blood pressure, body temperature, weight and the response to pain. Here we present structures of human NTSR1 in complex with the agonist JMV449 and the heterotrimeric G protein, at a resolution of 3 Å. We identify two conformations: a canonical-state complex that is similar to recently reported GPCR-G complexes (in which the nucleotide-binding pocket adopts more flexible conformations that may facilitate nucleotide exchange), and a non-canonical state in which the G protein is rotated by about 45 degrees relative to the receptor and exhibits a more rigid nucleotide-binding pocket. In the non-canonical state, NTSR1 exhibits features of both active and inactive conformations, which suggests that the structure may represent an intermediate form along the activation pathway of G proteins. This structural information, complemented by molecular dynamics simulations and functional studies, provides insights into the complex process of G-protein activation. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20181.map.gz emd_20181.map.gz | 49.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20181-v30.xml emd-20181-v30.xml emd-20181.xml emd-20181.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20181.png emd_20181.png | 93.9 KB | ||
| Filedesc metadata |  emd-20181.cif.gz emd-20181.cif.gz | 7.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20181 http://ftp.pdbj.org/pub/emdb/structures/EMD-20181 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20181 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20181 | HTTPS FTP |
-Related structure data
| Related structure data |  6osaMC  6os9C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20181.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20181.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | human neurotensin receptor 1 (hNTSR1) - Gi1 complex in non-canonical conformation (NC state) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : hNTSR1-Gi1 complex in non-canonical conformation (NC state)
| Entire | Name: hNTSR1-Gi1 complex in non-canonical conformation (NC state) |
|---|---|
| Components |
|
-Supramolecule #1: hNTSR1-Gi1 complex in non-canonical conformation (NC state)
| Supramolecule | Name: hNTSR1-Gi1 complex in non-canonical conformation (NC state) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Neurotensin receptor type 1
| Macromolecule | Name: Neurotensin receptor type 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 48.396734 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DYKDDDDAMG QPGNGSAFLL APNRSHAPDH DVENLYFQGQ RAQAGLEEAL LAPGFGNASG NASERVLAAP SSELDVNTDI YSKVLVTAV YLALFVVGTV GNTVTLFTLA RKKSLQSLQS TVHYHLGSLA LSDLLTLLLA MPVELYNFIW VHHPWAFGDA G CRGYYFLR ...String: DYKDDDDAMG QPGNGSAFLL APNRSHAPDH DVENLYFQGQ RAQAGLEEAL LAPGFGNASG NASERVLAAP SSELDVNTDI YSKVLVTAV YLALFVVGTV GNTVTLFTLA RKKSLQSLQS TVHYHLGSLA LSDLLTLLLA MPVELYNFIW VHHPWAFGDA G CRGYYFLR DACTYATALN VASLSVERYL AICHPFKAKT LMSRSRTKKF ISAIWLASAL LAVPMLFTMG EQNRSADGQH AG GLVCTPT IHTATVKVVI QVNTFMSFIF PMVVISVLNT IIANKLTVMV RQAAEQGQVC TVGGPGRVQA LRHGVRVLRA VVI AFVVCW LPYHVRRLMF CYISDEQWTP FLYDFYHYFY MVTNALFYVS STINPILYNL VSANFRHIFL ATLACLCPVW RRRR KRPAF SRKADSVSSN HTLSSNATRE TLYLEVLFQG UniProtKB: Neurotensin receptor type 1 |
-Macromolecule #2: JMV449
| Macromolecule | Name: JMV449 / type: protein_or_peptide / ID: 2 / Details: JMV449 from Tocris Bioscience. / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 762.979 Da |
| Sequence | String: KKPYIL |
-Macromolecule #3: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.415031 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGGQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHESM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCA TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.671102 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: PGSSGSELDQ LRQEAEQLKN QIRDARKACA DATLSQITNN IDPVGRIQMR TRRTLRGHLA KIYAMHWGTD SRLLVSASQD GKLIIWDSY TTNKVHAIPL RSSWVMTCAY APSGNYVACG GLDNICSIYN LKTREGNVRV SRELAGHTGY LSCCRFLDDN Q IVTSSGDT ...String: PGSSGSELDQ LRQEAEQLKN QIRDARKACA DATLSQITNN IDPVGRIQMR TRRTLRGHLA KIYAMHWGTD SRLLVSASQD GKLIIWDSY TTNKVHAIPL RSSWVMTCAY APSGNYVACG GLDNICSIYN LKTREGNVRV SRELAGHTGY LSCCRFLDDN Q IVTSSGDT TCALWDIETG QQTTTFTGHT GDVMSLSLAP DTRLFVSGAC DASAKLWDVR EGMCRQTFTG HESDINAICF FP NGNAFAT GSDDATCRLF DLRADQELMT YSHDNIICGI TSVSFSKSGR LLLAGYDDFN CNVWDALKAD RAGVLAGHDN RVS CLGVTD DGMAVATGSW DSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #5: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average exposure time: 10.0 sec. / Average electron dose: 75.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















