[English] 日本語
 Yorodumi
Yorodumi- PDB-1hpc: REFINED STRUCTURES AT 2 ANGSTROMS AND 2.2 ANGSTROMS OF THE TWO FO... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1hpc | ||||||
|---|---|---|---|---|---|---|---|
| Title | REFINED STRUCTURES AT 2 ANGSTROMS AND 2.2 ANGSTROMS OF THE TWO FORMS OF THE H-PROTEIN, A LIPOAMIDE-CONTAINING PROTEIN OF THE GLYCINE DECARBOXYLASE | ||||||
 Components Components | H PROTEIN OF THE GLYCINE CLEAVAGE SYSTEM | ||||||
 Keywords Keywords | TRANSIT PEPTIDE | ||||||
| Function / homology |  Function and homology information Function and homology informationglycine decarboxylation via glycine cleavage system / glycine cleavage complex / protein lipoylation / mitochondrial matrix Similarity search - Function | ||||||
| Biological species |  Pisum sativum (garden pea) Pisum sativum (garden pea) | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 2 Å X-RAY DIFFRACTION / Resolution: 2 Å | ||||||
 Authors Authors | Pares, S. / Cohen-Addad, C. / Sieker, L. / Neuburger, M. / Douce, R. | ||||||
 Citation Citation |  Journal: Acta Crystallogr.,Sect.D / Year: 1995 Journal: Acta Crystallogr.,Sect.D / Year: 1995Title: Refined structures at 2 and 2.2 A resolution of two forms of the H-protein, a lipoamide-containing protein of the glycine decarboxylase complex. Authors: Pares, S. / Cohen-Addad, C. / Sieker, L.C. / Neuburger, M. / Douce, R. #1:  Journal: Nat.Struct.Biol. / Year: 1995 Journal: Nat.Struct.Biol. / Year: 1995Title: The Lipoamide Arm in the Glycine Decarboxylase Complex is not Freely Swinging Authors: Cohen-Addad, C. / Pares, S. / Sieker, L. / Neuburger, M. / Douce, R. #2:  Journal: Proc.Natl.Acad.Sci.USA / Year: 1994 Journal: Proc.Natl.Acad.Sci.USA / Year: 1994Title: X-Ray Structure Determination at 2.6 Angstroms Resolution of a Lipoate-Containing Protein: The H-Protein of the Glycine Decarboxylase Complex from Pea Leaves Authors: Pares, S. / Cohen-Addad, C. / Sieker, L. / Neuburger, M. / Douce, R. #3:  Journal: J.Mol.Biol. / Year: 1991 Journal: J.Mol.Biol. / Year: 1991Title: Crystallographic Data for H-Protein from the Glycine Decarboxylase Complex Authors: Sieker, L. / Cohen-Addad, C. / Neuburger, M. / Douce, R. #4:  Journal: Biochem.J. / Year: 1990 Journal: Biochem.J. / Year: 1990Title: Cdna Cloning, Primary Structure and Gene Expression for H-Protein, a Component of the Glycine-Cleavage System (Glycine Decarboxylase) of Pea (Pisum Sativum) Leaf Mitochondria Authors: Macherel, D. / Lebrun, M. / Gagnon, J. / Neuburger, M. / Douce, R. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1hpc.cif.gz 1hpc.cif.gz | 74.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1hpc.ent.gz pdb1hpc.ent.gz | 55.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1hpc.json.gz 1hpc.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hp/1hpc https://data.pdbj.org/pub/pdb/validation_reports/hp/1hpc ftp://data.pdbj.org/pub/pdb/validation_reports/hp/1hpc ftp://data.pdbj.org/pub/pdb/validation_reports/hp/1hpc | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
| ||||||||
| Noncrystallographic symmetry (NCS) | NCS oper: (Code: given Matrix: (0.3895, 0.566, 0.726), Vector: Details | THE TRANSFORMATION PRESENTED ON *MTRIX* RECORDS BELOW WILL YIELD APPROXIMATE COORDINATES FOR CHAIN *B* WHEN APPLIED TO CHAIN *A*. | |
- Components
Components
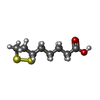
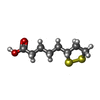
| #1: Protein | Mass: 13962.464 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Pisum sativum (garden pea) Pisum sativum (garden pea)References: UniProt: P16048, glycine dehydrogenase (aminomethyl-transferring) #2: Chemical | ChemComp-LPB / | #3: Chemical | ChemComp-LPA / | #4: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.32 Å3/Da / Density % sol: 46.99 % | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal | *PLUS Density % sol: 45 % | |||||||||||||||
| Crystal grow | *PLUS Temperature: 281 K / pH: 5.2 / Method: vapor diffusion | |||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Radiation | Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| Reflection | Num. obs: 17031 / % possible obs: 92 % |
| Reflection | *PLUS Highest resolution: 2 Å / Num. measured all: 87067 / Rmerge(I) obs: 0.081 |
| Reflection shell | *PLUS Highest resolution: 2 Å / Lowest resolution: 2.1 Å / % possible obs: 80 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 2→8 Å / σ(F): 2 /
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 23 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→8 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller












 PDBj
PDBj