[English] 日本語
 Yorodumi
Yorodumi- EMDB-19867: Cryo-EM Structure of a 1033 Scaffold Base DNA Origami Nanostructu... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of a 1033 Scaffold Base DNA Origami Nanostructure V4 and TBA | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | DNA Origami / Multilayer / Square lattice / 1033 scaffold / DNA / TBA | ||||||||||||
| Biological species | synthetic construct (others) | ||||||||||||
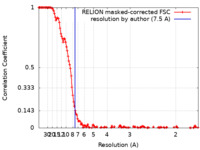
| Method | single particle reconstruction / cryo EM / Resolution: 7.5 Å | ||||||||||||
 Authors Authors | Ali K / Georg K / Volodymyr M / Johanna G / Maximilian NH / Lukas K / Simone C / Hendrik D | ||||||||||||
| Funding support | European Union,  Germany, 3 items Germany, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nano Lett / Year: 2024 Journal: Nano Lett / Year: 2024Title: Designing Rigid DNA Origami Templates for Molecular Visualization Using Cryo-EM. Authors: Ali Khoshouei / Georg Kempf / Volodymyr Mykhailiuk / Johanna Mariko Griessing / Maximilian Nicolas Honemann / Lukas Kater / Simone Cavadini / Hendrik Dietz /   Abstract: DNA origami, a method for constructing nanostructures from DNA, offers potential for diverse scientific and technological applications due to its ability to integrate various molecular ...DNA origami, a method for constructing nanostructures from DNA, offers potential for diverse scientific and technological applications due to its ability to integrate various molecular functionalities in a programmable manner. In this study, we examined the impact of internal crossover distribution and the compositional uniformity of staple strands on the structure of multilayer DNA origami using cryogenic electron microscopy (cryo-EM) single-particle analysis. A refined DNA object was utilized as an alignment framework in a host-guest model, where we successfully resolved an 8 kDa thrombin binding aptamer (TBA) linked to the host object. Our results broaden the spectrum of DNA in structural applications. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19867.map.gz emd_19867.map.gz | 191.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19867-v30.xml emd-19867-v30.xml emd-19867.xml emd-19867.xml | 22.9 KB 22.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19867_fsc.xml emd_19867_fsc.xml | 14.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_19867.png emd_19867.png | 58 KB | ||
| Masks |  emd_19867_msk_1.map emd_19867_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19867.cif.gz emd-19867.cif.gz | 5 KB | ||
| Others |  emd_19867_additional_1.map.gz emd_19867_additional_1.map.gz emd_19867_additional_2.map.gz emd_19867_additional_2.map.gz emd_19867_additional_3.map.gz emd_19867_additional_3.map.gz emd_19867_half_map_1.map.gz emd_19867_half_map_1.map.gz emd_19867_half_map_2.map.gz emd_19867_half_map_2.map.gz | 216.2 MB 216.3 MB 216.3 MB 193.3 MB 193.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19867 http://ftp.pdbj.org/pub/emdb/structures/EMD-19867 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19867 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19867 | HTTPS FTP |
-Validation report
| Summary document |  emd_19867_validation.pdf.gz emd_19867_validation.pdf.gz | 790.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19867_full_validation.pdf.gz emd_19867_full_validation.pdf.gz | 790.4 KB | Display | |
| Data in XML |  emd_19867_validation.xml.gz emd_19867_validation.xml.gz | 21.7 KB | Display | |
| Data in CIF |  emd_19867_validation.cif.gz emd_19867_validation.cif.gz | 28.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19867 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19867 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19867 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19867 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19867.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19867.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
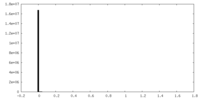
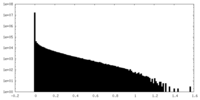
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19867_msk_1.map emd_19867_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
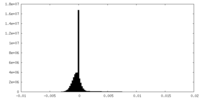
| Projections & Slices |
| ||||||||||||
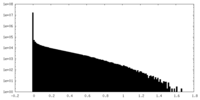
| Density Histograms |
-Additional map: Body1
| File | emd_19867_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Body1 | ||||||||||||
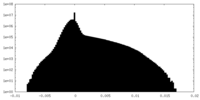
| Projections & Slices |
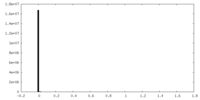
| ||||||||||||
| Density Histograms |
-Additional map: Body2
| File | emd_19867_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Body2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
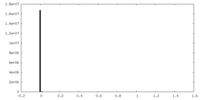
| Density Histograms |
-Additional map: Body3
| File | emd_19867_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Body3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_19867_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
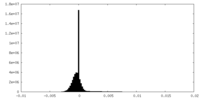
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_19867_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM Structure of a 1033 Scaffold Base DNA Origami Nanostructu...
| Entire | Name: Cryo-EM Structure of a 1033 Scaffold Base DNA Origami Nanostructure V4 and TBA |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM Structure of a 1033 Scaffold Base DNA Origami Nanostructu...
| Supramolecule | Name: Cryo-EM Structure of a 1033 Scaffold Base DNA Origami Nanostructure V4 and TBA type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: DNA (42-MER)
| Macromolecule | Name: DNA (42-MER) / type: dna / ID: 1 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 13.034333 KDa |
| Sequence | String: (DA)(DC)(DT)(DT)(DC)(DC)(DA)(DG)(DT)(DC) (DC)(DG)(DT)(DG)(DG)(DT)(DA)(DG)(DG)(DG) (DC)(DA)(DG)(DG)(DT)(DT)(DG)(DG)(DG) (DG)(DT)(DG)(DA)(DC)(DC)(DG)(DC)(DT)(DA) (DT) (DA)(DT) |
-Macromolecule #2: DNA (5'-D(P*AP*TP*AP*TP*AP*GP*CP*GP*TP*GP*GP*AP*AP*GP*T)-3')
| Macromolecule | Name: DNA (5'-D(P*AP*TP*AP*TP*AP*GP*CP*GP*TP*GP*GP*AP*AP*GP*T)-3') type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 4.673059 KDa |
| Sequence | String: (DA)(DT)(DA)(DT)(DA)(DG)(DC)(DG)(DT)(DG) (DG)(DA)(DA)(DG)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 75000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)