+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
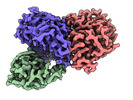
| タイトル | Cryo-EM structure of CDK2-cyclin A in complex with CDC25A | |||||||||
 マップデータ マップデータ | ||||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | cell-cycle / CDK / phosphatase / cancer / CELL CYCLE | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報positive regulation of G2/MI transition of meiotic cell cycle / cell cycle G1/S phase transition / mitotic cell cycle phase transition / Transcription of E2F targets under negative control by DREAM complex / Polo-like kinase mediated events / cyclin-dependent protein serine/threonine kinase regulator activity / Deregulated CDK5 triggers multiple neurodegenerative pathways in Alzheimer's disease models / regulation of cyclin-dependent protein serine/threonine kinase activity / cyclin A1-CDK2 complex / cyclin E2-CDK2 complex ...positive regulation of G2/MI transition of meiotic cell cycle / cell cycle G1/S phase transition / mitotic cell cycle phase transition / Transcription of E2F targets under negative control by DREAM complex / Polo-like kinase mediated events / cyclin-dependent protein serine/threonine kinase regulator activity / Deregulated CDK5 triggers multiple neurodegenerative pathways in Alzheimer's disease models / regulation of cyclin-dependent protein serine/threonine kinase activity / cyclin A1-CDK2 complex / cyclin E2-CDK2 complex / cyclin E1-CDK2 complex / cyclin A2-CDK2 complex / positive regulation of DNA-templated DNA replication initiation / G2 Phase / cyclin-dependent protein kinase activity / Y chromosome / Phosphorylation of proteins involved in G1/S transition by active Cyclin E:Cdk2 complexes / positive regulation of heterochromatin formation / p53-Dependent G1 DNA Damage Response / X chromosome / PTK6 Regulates Cell Cycle / regulation of anaphase-promoting complex-dependent catabolic process / regulation of DNA replication / Defective binding of RB1 mutants to E2F1,(E2F2, E2F3) / centriole replication / Regulation of APC/C activators between G1/S and early anaphase / telomere maintenance in response to DNA damage / centrosome duplication / phosphoprotein phosphatase activity / G0 and Early G1 / Telomere Extension By Telomerase / cellular response to nitric oxide / Activation of the pre-replicative complex / cyclin-dependent kinase / cyclin-dependent protein serine/threonine kinase activity / TP53 Regulates Transcription of Genes Involved in G1 Cell Cycle Arrest / Regulation of MITF-M-dependent genes involved in cell cycle and proliferation / Activation of ATR in response to replication stress / Cyclin E associated events during G1/S transition / Cajal body / Cyclin A/B1/B2 associated events during G2/M transition / cyclin-dependent protein kinase holoenzyme complex / mitotic G1 DNA damage checkpoint signaling / Cyclin A:Cdk2-associated events at S phase entry / condensed chromosome / regulation of G2/M transition of mitotic cell cycle / regulation of mitotic cell cycle / positive regulation of G2/M transition of mitotic cell cycle / post-translational protein modification / cyclin binding / protein-tyrosine-phosphatase / protein tyrosine phosphatase activity / positive regulation of DNA replication / male germ cell nucleus / meiotic cell cycle / Ubiquitin Mediated Degradation of Phosphorylated Cdc25A / response to radiation / potassium ion transport / DNA Damage/Telomere Stress Induced Senescence / CDK-mediated phosphorylation and removal of Cdc6 / Meiotic recombination / SCF(Skp2)-mediated degradation of p27/p21 / Orc1 removal from chromatin / Transcriptional regulation of granulopoiesis / G1/S transition of mitotic cell cycle / cellular response to UV / Cyclin D associated events in G1 / G2/M transition of mitotic cell cycle / cellular senescence / Regulation of TP53 Degradation / nuclear envelope / protein-folding chaperone binding / Factors involved in megakaryocyte development and platelet production / Processing of DNA double-strand break ends / Senescence-Associated Secretory Phenotype (SASP) / peptidyl-serine phosphorylation / regulation of gene expression / Regulation of TP53 Activity through Phosphorylation / transcription regulator complex / Ras protein signal transduction / DNA replication / cell population proliferation / chromosome, telomeric region / Ub-specific processing proteases / endosome / chromatin remodeling / protein phosphorylation / protein domain specific binding / cell division / protein serine kinase activity / DNA repair / protein serine/threonine kinase activity / centrosome / DNA-templated transcription / positive regulation of cell population proliferation / protein kinase binding / positive regulation of DNA-templated transcription / negative regulation of transcription by RNA polymerase II / magnesium ion binding / signal transduction 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) / Homo sapiens (ヒト) /  | |||||||||
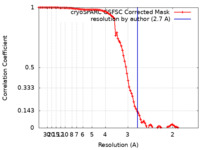
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.7 Å | |||||||||
 データ登録者 データ登録者 | Rowland RJ / Noble MEM / Endicott JA | |||||||||
| 資金援助 |  英国, 2件 英国, 2件
| |||||||||
 引用 引用 |  ジャーナル: Nat Commun / 年: 2024 ジャーナル: Nat Commun / 年: 2024タイトル: Cryo-EM structure of the CDK2-cyclin A-CDC25A complex. 著者: Rhianna J Rowland / Svitlana Korolchuk / Marco Salamina / Natalie J Tatum / James R Ault / Sam Hart / Johan P Turkenburg / James N Blaza / Martin E M Noble / Jane A Endicott /  要旨: The cell division cycle 25 phosphatases CDC25A, B and C regulate cell cycle transitions by dephosphorylating residues in the conserved glycine-rich loop of CDKs to activate their activity. Here, we ...The cell division cycle 25 phosphatases CDC25A, B and C regulate cell cycle transitions by dephosphorylating residues in the conserved glycine-rich loop of CDKs to activate their activity. Here, we present the cryo-EM structure of CDK2-cyclin A in complex with CDC25A at 2.7 Å resolution, providing a detailed structural analysis of the overall complex architecture and key protein-protein interactions that underpin this 86 kDa complex. We further identify a CDC25A C-terminal helix that is critical for complex formation. Sequence conservation analysis suggests CDK1/2-cyclin A, CDK1-cyclin B and CDK2/3-cyclin E are suitable binding partners for CDC25A, whilst CDK4/6-cyclin D complexes appear unlikely substrates. A comparative structural analysis of CDK-containing complexes also confirms the functional importance of the conserved CDK1/2 GDSEID motif. This structure improves our understanding of the roles of CDC25 phosphatases in CDK regulation and may inform the development of CDC25-targeting anticancer strategies. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_19408.map.gz emd_19408.map.gz | 85.8 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-19408-v30.xml emd-19408-v30.xml emd-19408.xml emd-19408.xml | 21.6 KB 21.6 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_19408_fsc.xml emd_19408_fsc.xml | 9.5 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_19408.png emd_19408.png | 122.1 KB | ||
| マスクデータ |  emd_19408_msk_1.map emd_19408_msk_1.map | 91.1 MB |  マスクマップ マスクマップ | |
| Filedesc metadata |  emd-19408.cif.gz emd-19408.cif.gz | 7 KB | ||
| その他 |  emd_19408_half_map_1.map.gz emd_19408_half_map_1.map.gz emd_19408_half_map_2.map.gz emd_19408_half_map_2.map.gz | 84.6 MB 84.6 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19408 http://ftp.pdbj.org/pub/emdb/structures/EMD-19408 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19408 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19408 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_19408_validation.pdf.gz emd_19408_validation.pdf.gz | 805.7 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_19408_full_validation.pdf.gz emd_19408_full_validation.pdf.gz | 805.3 KB | 表示 | |
| XML形式データ |  emd_19408_validation.xml.gz emd_19408_validation.xml.gz | 17.6 KB | 表示 | |
| CIF形式データ |  emd_19408_validation.cif.gz emd_19408_validation.cif.gz | 22.6 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19408 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19408 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19408 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19408 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8rozMC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_19408.map.gz / 形式: CCP4 / 大きさ: 91.1 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_19408.map.gz / 形式: CCP4 / 大きさ: 91.1 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-マスク #1
| ファイル |  emd_19408_msk_1.map emd_19408_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: #2
| ファイル | emd_19408_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: #1
| ファイル | emd_19408_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Trimeric complex of CDK2-cyclin A-CDC25A
| 全体 | 名称: Trimeric complex of CDK2-cyclin A-CDC25A |
|---|---|
| 要素 |
|
-超分子 #1: Trimeric complex of CDK2-cyclin A-CDC25A
| 超分子 | 名称: Trimeric complex of CDK2-cyclin A-CDC25A / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: all |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 86 KDa |
-分子 #1: Cyclin-dependent kinase 2
| 分子 | 名称: Cyclin-dependent kinase 2 / タイプ: protein_or_peptide / ID: 1 詳細: Human cyclin dependant protein kinase 2 (CDK2) phosphorylated at Tyr15 and Thr160 コピー数: 1 / 光学異性体: LEVO / EC番号: cyclin-dependent kinase |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 34.136449 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MENFQKVEKI GEGT(PTR)GVVYK ARNKLTGEVV ALKKIRLDTE TEGVPSTAIR EISLLKELNH PNIVKLLDVI HTENKL YLV FEFLHQDLKK FMDASALTGI PLPLIKSYLF QLLQGLAFCH SHRVLHRDLK PQNLLINTEG AIKLADFGLA RAFGVPV RT Y(TPO) ...文字列: MENFQKVEKI GEGT(PTR)GVVYK ARNKLTGEVV ALKKIRLDTE TEGVPSTAIR EISLLKELNH PNIVKLLDVI HTENKL YLV FEFLHQDLKK FMDASALTGI PLPLIKSYLF QLLQGLAFCH SHRVLHRDLK PQNLLINTEG AIKLADFGLA RAFGVPV RT Y(TPO)HEVVTLWY RAPEILLGCK YYSTAVDIWS LGCIFAEMVT RRALFPGDSE IDQLFRIFRT LGTPDEVVWP GVTS MPDYK PSFPKWARQD FSKVVPPLDE DGRSLLSQML HYDPNKRISA KAALAHPFFQ DVTKPVPHLR L UniProtKB: Cyclin-dependent kinase 2 |
-分子 #2: Cyclin-A2
| 分子 | 名称: Cyclin-A2 / タイプ: protein_or_peptide / ID: 2 / 詳細: Bovine cyclin A2 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 30.119799 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MGVNEVPDYH EDIHTYLREM EVKCKPKVGY MKKQPDITNS MRAILVDWLV EVGEEYKLQN ETLHLAVNYI DRFLSSMSVL RGKLQLVGT AAMLLASKFE EIYPPEVAEF VYITDDTYTK KQVLRMEHLV LKVLAFDLAA PTINQFLTQY FLHQQPANCK V ESLAMFLG ...文字列: MGVNEVPDYH EDIHTYLREM EVKCKPKVGY MKKQPDITNS MRAILVDWLV EVGEEYKLQN ETLHLAVNYI DRFLSSMSVL RGKLQLVGT AAMLLASKFE EIYPPEVAEF VYITDDTYTK KQVLRMEHLV LKVLAFDLAA PTINQFLTQY FLHQQPANCK V ESLAMFLG ELSLIDADPY LKYLPSVIAA AAFHLALYTV TGQSWPESLV QKTGYTLETL KPCLLDLHQT YLRAPQHAQQ SI REKYKNS KYHGVSLLNP PETLNV UniProtKB: Cyclin-A2 |
-分子 #3: M-phase inducer phosphatase 1
| 分子 | 名称: M-phase inducer phosphatase 1 / タイプ: protein_or_peptide / ID: 3 詳細: M-phase inducer phosphatase 1 (CDC25A) catalytic domain コピー数: 1 / 光学異性体: LEVO / EC番号: protein-tyrosine-phosphatase |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 22.533123 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: SMVIGDFSKG YLFHTVAGKH QDLKYISPEI MASVLNGKFA NLIKEFVIID CRYPYEYEGG HIKGAVNLHM EEEVEDFLLK KPIVPTDGK RVIVVFHSEF SSERGPRMCR YVRERDRLGN EYPKLHYPEL YVLKGGYKEF FMKCQSYCEP PSYRPMHHED F KEDLKKFR ...文字列: SMVIGDFSKG YLFHTVAGKH QDLKYISPEI MASVLNGKFA NLIKEFVIID CRYPYEYEGG HIKGAVNLHM EEEVEDFLLK KPIVPTDGK RVIVVFHSEF SSERGPRMCR YVRERDRLGN EYPKLHYPEL YVLKGGYKEF FMKCQSYCEP PSYRPMHHED F KEDLKKFR TKSRTWAGEK SKREMYSRLK KL UniProtKB: M-phase inducer phosphatase 1 |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 1.8 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 緩衝液 | pH: 7.4 構成要素:
詳細: 50 mM HEPES, 200 mM NaCl, 1 mM DTT, pH 7.4 | ||||||||||||
| グリッド | モデル: Quantifoil R1.2/1.3 / 材質: COPPER / メッシュ: 200 / 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: HOLEY / 前処理 - タイプ: GLOW DISCHARGE / 前処理 - 時間: 60 sec. / 前処理 - 雰囲気: AIR / 前処理 - 気圧: 0.026000000000000002 kPa 詳細: Grids were glow discharged for 1min at 20 mAmp/0.26 mBar | ||||||||||||
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 80 % / チャンバー内温度: 277 K / 装置: FEI VITROBOT MARK IV |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | TFS KRIOS |
|---|---|
| 特殊光学系 | エネルギーフィルター - 名称: GIF Bioquantum / エネルギーフィルター - スリット幅: 20 eV |
| 撮影 | フィルム・検出器のモデル: GATAN K3 BIOQUANTUM (6k x 4k) 平均露光時間: 1.92 sec. / 平均電子線量: 50.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | C2レンズ絞り径: 100.0 µm / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最大 デフォーカス(公称値): 2.0 µm / 最小 デフォーカス(公称値): 0.6 µm / 倍率(公称値): 150000 |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
+ 画像解析
画像解析
-原子モデル構築 1
| 初期モデル | Chain - Source name: Other / Chain - Initial model type: in silico model / 詳細: ModelAngelo |
|---|---|
| 詳細 | A model was generated using ModelAngelo. Initial local fitting was performed in ChimerX followed by real space refinement in Phenix and manual fitting in coot. |
| 精密化 | 空間: REAL / プロトコル: AB INITIO MODEL / 温度因子: 144 |
| 得られたモデル |  PDB-8roz: |
 ムービー
ムービー コントローラー
コントローラー


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)