+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Human Replication protein A (RPA; trimeric core) - ssDNA complex | |||||||||||||||||||||||||||
 マップデータ マップデータ | ||||||||||||||||||||||||||||
 試料 試料 |
| |||||||||||||||||||||||||||
 キーワード キーワード | ssDNA binding protein / DNA damage repair / Single-strand annealing / DNA BINDING PROTEIN | |||||||||||||||||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報protein localization to chromosome / DNA replication factor A complex / chromatin-protein adaptor activity / single-stranded telomeric DNA binding / regulation of DNA damage checkpoint / Removal of the Flap Intermediate / Mismatch repair (MMR) directed by MSH2:MSH3 (MutSbeta) / Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) / protein localization to site of double-strand break / Removal of the Flap Intermediate from the C-strand ...protein localization to chromosome / DNA replication factor A complex / chromatin-protein adaptor activity / single-stranded telomeric DNA binding / regulation of DNA damage checkpoint / Removal of the Flap Intermediate / Mismatch repair (MMR) directed by MSH2:MSH3 (MutSbeta) / Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) / protein localization to site of double-strand break / Removal of the Flap Intermediate from the C-strand / G-rich strand telomeric DNA binding / HDR through Single Strand Annealing (SSA) / Impaired BRCA2 binding to RAD51 / regulation of double-strand break repair via homologous recombination / telomeric DNA binding / site of DNA damage / Presynaptic phase of homologous DNA pairing and strand exchange / telomere maintenance via telomerase / PCNA-Dependent Long Patch Base Excision Repair / HSF1 activation / Regulation of HSF1-mediated heat shock response / Activation of the pre-replicative complex / mismatch repair / Activation of ATR in response to replication stress / SUMOylation of DNA damage response and repair proteins / mitotic G1 DNA damage checkpoint signaling / regulation of mitotic cell cycle / telomere maintenance / Translesion synthesis by REV1 / Translesion synthesis by POLK / Translesion synthesis by POLI / Gap-filling DNA repair synthesis and ligation in GG-NER / meiotic cell cycle / nucleotide-excision repair / Fanconi Anemia Pathway / Recognition of DNA damage by PCNA-containing replication complex / Termination of translesion DNA synthesis / double-strand break repair via homologous recombination / Translesion Synthesis by POLH / base-excision repair / G2/M DNA damage checkpoint / HDR through Homologous Recombination (HRR) / PML body / Dual Incision in GG-NER / DNA-templated DNA replication / Meiotic recombination / Formation of Incision Complex in GG-NER / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / site of double-strand break / single-stranded DNA binding / regulation of cell population proliferation / Processing of DNA double-strand break ends / protein phosphatase binding / DNA recombination / Regulation of TP53 Activity through Phosphorylation / DNA replication / damaged DNA binding / chromosome, telomeric region / nuclear body / DNA repair / DNA damage response / ubiquitin protein ligase binding / chromatin / enzyme binding / nucleoplasm / nucleus / metal ion binding 類似検索 - 分子機能 | |||||||||||||||||||||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||||||||||||||||||||
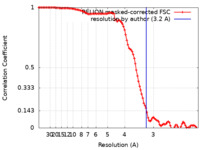
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.2 Å | |||||||||||||||||||||||||||
 データ登録者 データ登録者 | Liang CC / West SC | |||||||||||||||||||||||||||
| 資金援助 |  英国, European Union, 英国, European Union,  スイス, 8件 スイス, 8件
| |||||||||||||||||||||||||||
 引用 引用 |  ジャーナル: Nature / 年: 2024 ジャーナル: Nature / 年: 2024タイトル: Mechanism of single-stranded DNA annealing by RAD52-RPA complex. 著者: Chih-Chao Liang / Luke A Greenhough / Laura Masino / Sarah Maslen / Ilirjana Bajrami / Marcel Tuppi / Mark Skehel / Ian A Taylor / Stephen C West /  要旨: RAD52 is important for the repair of DNA double-stranded breaks, mitotic DNA synthesis and alternative telomere length maintenance. Central to these functions, RAD52 promotes the annealing of ...RAD52 is important for the repair of DNA double-stranded breaks, mitotic DNA synthesis and alternative telomere length maintenance. Central to these functions, RAD52 promotes the annealing of complementary single-stranded DNA (ssDNA) and provides an alternative to BRCA2/RAD51-dependent homologous recombination repair. Inactivation of RAD52 in homologous-recombination-deficient BRCA1- or BRCA2-defective cells is synthetically lethal, and aberrant expression of RAD52 is associated with poor cancer prognosis. As a consequence, RAD52 is an attractive therapeutic target against homologous-recombination-deficient breast, ovarian and prostate cancers. Here we describe the structure of RAD52 and define the mechanism of annealing. As reported previously, RAD52 forms undecameric (11-subunit) ring structures, but these rings do not represent the active form of the enzyme. Instead, cryo-electron microscopy and biochemical analyses revealed that ssDNA annealing is driven by RAD52 open rings in association with replication protein-A (RPA). Atomic models of the RAD52-ssDNA complex show that ssDNA sits in a positively charged channel around the ring. Annealing is driven by the RAD52 N-terminal domains, whereas the C-terminal regions modulate the open-ring conformation and RPA interaction. RPA associates with RAD52 at the site of ring opening with critical interactions occurring between the RPA-interacting domain of RAD52 and the winged helix domain of RPA2. Our studies provide structural snapshots throughout the annealing process and define the molecular mechanism of ssDNA annealing by the RAD52-RPA complex. | |||||||||||||||||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_19255.map.gz emd_19255.map.gz | 6.7 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-19255-v30.xml emd-19255-v30.xml emd-19255.xml emd-19255.xml | 21.4 KB 21.4 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_19255_fsc.xml emd_19255_fsc.xml | 10.6 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_19255.png emd_19255.png | 55.2 KB | ||
| Filedesc metadata |  emd-19255.cif.gz emd-19255.cif.gz | 6.8 KB | ||
| その他 |  emd_19255_half_map_1.map.gz emd_19255_half_map_1.map.gz emd_19255_half_map_2.map.gz emd_19255_half_map_2.map.gz | 80.8 MB 80.8 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19255 http://ftp.pdbj.org/pub/emdb/structures/EMD-19255 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19255 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19255 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_19255_validation.pdf.gz emd_19255_validation.pdf.gz | 542.8 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_19255_full_validation.pdf.gz emd_19255_full_validation.pdf.gz | 542.4 KB | 表示 | |
| XML形式データ |  emd_19255_validation.xml.gz emd_19255_validation.xml.gz | 17.6 KB | 表示 | |
| CIF形式データ |  emd_19255_validation.cif.gz emd_19255_validation.cif.gz | 23.2 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19255 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19255 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19255 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19255 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_19255.map.gz / 形式: CCP4 / 大きさ: 103 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_19255.map.gz / 形式: CCP4 / 大きさ: 103 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-ハーフマップ: #1
| ファイル | emd_19255_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: #2
| ファイル | emd_19255_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Open ring conformation of human RAD52 in complex with ssDNA
| 全体 | 名称: Open ring conformation of human RAD52 in complex with ssDNA |
|---|---|
| 要素 |
|
-超分子 #1: Open ring conformation of human RAD52 in complex with ssDNA
| 超分子 | 名称: Open ring conformation of human RAD52 in complex with ssDNA タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: all 詳細: Recombinant RAD52 purified from E. coli, and the open ring conformation was separated by cation exchange. The RAD52-ssDNA complex was reconstituted in vitro. |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-分子 #1: Replication protein A 70 kDa DNA-binding subunit, N-terminally pr...
| 分子 | 名称: Replication protein A 70 kDa DNA-binding subunit, N-terminally processed タイプ: protein_or_peptide / ID: 1 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 68.212977 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MVGQLSEGAI AAIMQKGDTN IKPILQVINI RPITTGNSPP RYRLLMSDGL NTLSSFMLAT QLNPLVEEEQ LSSNCVCQIH RFIVNTLKD GRRVVILMEL EVLKSAEAVG VKIGNPVPYN EGLGQPQVAP PAPAASPAAS SRPQPQNGSS GMGSTVSKAY G ASKTFGKA ...文字列: MVGQLSEGAI AAIMQKGDTN IKPILQVINI RPITTGNSPP RYRLLMSDGL NTLSSFMLAT QLNPLVEEEQ LSSNCVCQIH RFIVNTLKD GRRVVILMEL EVLKSAEAVG VKIGNPVPYN EGLGQPQVAP PAPAASPAAS SRPQPQNGSS GMGSTVSKAY G ASKTFGKA AGPSLSHTSG GTQSKVVPIA SLTPYQSKWT ICARVTNKSQ IRTWSNSRGE GKLFSLELVD ESGEIRATAF NE QVDKFFP LIEVNKVYYF SKGTLKIANK QFTAVKNDYE MTFNNETSVM PCEDDHHLPT VQFDFTGIDD LENKSKDSLV DII GICKSY EDATKITVRS NNREVAKRNI YLMDTSGKVV TATLWGEDAD KFDGSRQPVL AIKGARVSDF GGRSLSVLSS STII ANPDI PEAYKLRGWF DAEGQALDGV SISDLKSGGV GGSNTNWKTL YEVKSENLGQ GDKPDYFSSV ATVVYLRKEN CMYQA CPTQ DCNKKVIDQQ NGLYRCEKCD TEFPNFKYRM ILSVNIADFQ ENQWVTCFQE SAEAILGQNA AYLGELKDKN EQAFEE VFQ NANFRSFIFR VRVKVETYND ESRIKATVMD VKPVDYREYG RRLVMSIRRS ALM UniProtKB: Replication protein A 70 kDa DNA-binding subunit |
-分子 #2: Replication protein A 32 kDa subunit
| 分子 | 名称: Replication protein A 32 kDa subunit / タイプ: protein_or_peptide / ID: 2 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 29.276795 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MWNSGFESYG SSSYGGAGGY TQSPGGFGSP APSQAEKKSR ARAQHIVPCT ISQLLSATLV DEVFRIGNVE ISQVTIVGII RHAEKAPTN IVYKIDDMTA APMDVRQWVD TDDTSSENTV VPPETYVKVA GHLRSFQNKK SLVAFKIMPL EDMNEFTTHI L EVINAHMV ...文字列: MWNSGFESYG SSSYGGAGGY TQSPGGFGSP APSQAEKKSR ARAQHIVPCT ISQLLSATLV DEVFRIGNVE ISQVTIVGII RHAEKAPTN IVYKIDDMTA APMDVRQWVD TDDTSSENTV VPPETYVKVA GHLRSFQNKK SLVAFKIMPL EDMNEFTTHI L EVINAHMV LSKANSQPSA GRAPISNPGM SEAGNFGGNS FMPANGLTVA QNQVLNLIKA CPRPEGLNFQ DLKNQLKHMS VS SIKQAVD FLSNEGHIYS TVDDDHFKST DAE UniProtKB: Replication protein A 32 kDa subunit |
-分子 #3: Replication protein A 14 kDa subunit
| 分子 | 名称: Replication protein A 14 kDa subunit / タイプ: protein_or_peptide / ID: 3 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 13.583714 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MVDMMDLPRS RINAGMLAQF IDKPVCFVGR LEKIHPTGKM FILSDGEGKN GTIELMEPLD EEISGIVEVV GRVTAKATIL CTSYVQFKE DSHPFDLGLY NEAVKIIHDF PQFYPLGIVQ HD UniProtKB: Replication protein A 14 kDa subunit |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 0.15 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 緩衝液 | pH: 8 構成要素:
| ||||||||||||||||||
| グリッド | モデル: UltrAuFoil R2/2 / 材質: GOLD / メッシュ: 200 | ||||||||||||||||||
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 95 % / チャンバー内温度: 277.15 K / 装置: FEI VITROBOT MARK IV |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K2 QUANTUM (4k x 4k) 検出モード: COUNTING / 平均電子線量: 46.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | C2レンズ絞り径: 50.0 µm / 照射モード: SPOT SCAN / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最大 デフォーカス(公称値): 3.5 µm / 最小 デフォーカス(公称値): 1.5 µm / 倍率(公称値): 130000 |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
+ 画像解析
画像解析
-原子モデル構築 1
| 初期モデル | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| 精密化 | 空間: REAL / プロトコル: RIGID BODY FIT |
| 得られたモデル |  PDB-8rk2: |
 ムービー
ムービー コントローラー
コントローラー























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)