[English] 日本語
 Yorodumi
Yorodumi- EMDB-18861: CryoEM structure of the asymmetric Pho90 dimer from yeast without... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of the asymmetric Pho90 dimer from yeast without substrates. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Phosphate transporter / Plasma membrane protein / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of phosphate transmembrane transport / phosphate transmembrane transporter activity / polyphosphate metabolic process / phosphate ion transport / cell periphery / transmembrane transport / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
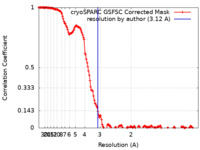
| Method | single particle reconstruction / cryo EM / Resolution: 3.12 Å | |||||||||
 Authors Authors | Schneider S / Kuehlbrandt W / Yildiz O | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2024 Journal: Structure / Year: 2024Title: Complementary structures of the yeast phosphate transporter Pho90 provide insights into its transport mechanism. Authors: Simon Schneider / Werner Kühlbrandt / Özkan Yildiz /  Abstract: Phosphate homeostasis is essential for all living organisms. Low-affinity phosphate transporters are involved in phosphate import and regulation in a range of eukaryotic organisms. We have determined ...Phosphate homeostasis is essential for all living organisms. Low-affinity phosphate transporters are involved in phosphate import and regulation in a range of eukaryotic organisms. We have determined the structures of the Saccharomyces cerevisiae phosphate importer Pho90 by electron cryomicroscopy in two complementary states at 2.3 and 3.1 Å resolution. The symmetrical, outward-open structure in the presence of phosphate indicates bound substrate ions in the binding pocket. In the absence of phosphate, Pho90 assumes an asymmetric structure with one monomer facing inward and one monomer facing outward, providing insights into the transport mechanism. The Pho90 transport domain binds phosphate ions on one side of the membrane, then flips to the other side where the substrate is released. Together with functional experiments, these complementary structures illustrate the transport mechanism of eukaryotic low-affinity phosphate transporters. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18861.map.gz emd_18861.map.gz | 323.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18861-v30.xml emd-18861-v30.xml emd-18861.xml emd-18861.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18861_fsc.xml emd_18861_fsc.xml | 14.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_18861.png emd_18861.png | 101.2 KB | ||
| Filedesc metadata |  emd-18861.cif.gz emd-18861.cif.gz | 6 KB | ||
| Others |  emd_18861_additional_1.map.gz emd_18861_additional_1.map.gz emd_18861_half_map_1.map.gz emd_18861_half_map_1.map.gz emd_18861_half_map_2.map.gz emd_18861_half_map_2.map.gz | 169.4 MB 318.1 MB 318.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18861 http://ftp.pdbj.org/pub/emdb/structures/EMD-18861 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18861 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18861 | HTTPS FTP |
-Validation report
| Summary document |  emd_18861_validation.pdf.gz emd_18861_validation.pdf.gz | 1012.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18861_full_validation.pdf.gz emd_18861_full_validation.pdf.gz | 1012.3 KB | Display | |
| Data in XML |  emd_18861_validation.xml.gz emd_18861_validation.xml.gz | 24.3 KB | Display | |
| Data in CIF |  emd_18861_validation.cif.gz emd_18861_validation.cif.gz | 31.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18861 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18861 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18861 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18861 | HTTPS FTP |
-Related structure data
| Related structure data |  8r35MC  8r33C  8r34C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18861.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18861.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.573 Å | ||||||||||||||||||||||||||||||||||||
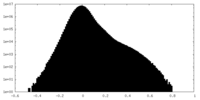
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_18861_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
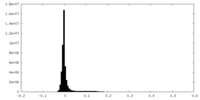
| Density Histograms |
-Half map: #2
| File | emd_18861_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
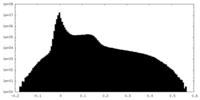
| Density Histograms |
-Half map: #1
| File | emd_18861_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
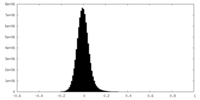
| Density Histograms |
- Sample components
Sample components
-Entire : Dimeric ScPho90
| Entire | Name: Dimeric ScPho90 |
|---|---|
| Components |
|
-Supramolecule #1: Dimeric ScPho90
| Supramolecule | Name: Dimeric ScPho90 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Low-affinity phosphate transporter PHO90
| Macromolecule | Name: Low-affinity phosphate transporter PHO90 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 97.786219 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRFSHFLKYN AVPEWQNHYM DYSELKNLIY TLQTDELQVG DNEEGFGAGK SSNITDRFKN KFSFKNAKED TSSGMNKDAG IVEETIELR ELPTAQTVAA KPSPFRRMKE KIFYKRRSSS ASSVSSTANE NLQLDTYDTF VGDLTAEKQK VDDFYKRTEA K FYDKFDAL ...String: MRFSHFLKYN AVPEWQNHYM DYSELKNLIY TLQTDELQVG DNEEGFGAGK SSNITDRFKN KFSFKNAKED TSSGMNKDAG IVEETIELR ELPTAQTVAA KPSPFRRMKE KIFYKRRSSS ASSVSSTANE NLQLDTYDTF VGDLTAEKQK VDDFYKRTEA K FYDKFDAL VKDLKKIGVI EYDIDDDTLF NEPIASTNDE VPPLDLDDDE DDDEFYDDQS NIEDNTALLH HSQYNIKSQK KS LLKKSIV NLYIDLCQLK SFIELNRIGF AKITKKSDKV LHLNTRTELI ESEQFFKDTY AFQAETIELL NSKISQLVTF YAR ITDRPH NISHSKQELK SYLHDHIVWE RSNTWKDMLG LLSQADELTP KETEYNANKL VGKLDLEYYR WPLPRPINLK FTSI NNVAL PKLFFTKKAY KIYFIILVTG LLLGIKTFND AAQHRCMALV ECVAFLWASE AIPLHITAFL VPLLVVLFKV LKTSD GAIM SAASASSEIL AAMWSSTIMI LLAGFTLGEV LAQYNIAKVL ASWLLAFAGC KPRNVLLMAM CVVFFLSMWI SNVAAP VLT YSLLSPLLDA MDADSPFAQA LVLGVALAAN IGGMSSPISS PQNIISMSYL KPYGIGWGQF FAVALPSGIL AMLLVWI LL FTTFKMNKTK LEKFKPIKTK FTVKQYYIIT VTVATILLWC VESQIEGAFG SSGQIAIIPI VLFFGTGLLS TQDLNAFP W SIVILAMGGI ALGKAVSSSG LLSTIAKALQ KKIENDGVFA ILCIFGILML VVGTFVSHTV SAIIIIPLVQ EVGDKLGNP KAAPILVFGC ALLSSCGMGL ASSGFPNVTA ISKVDRKGDR YLSVMTFLTR GVPASILAFL CVITLGYGIM ASVVKGNATS A UniProtKB: Low-affinity phosphate transporter PHO90 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.0 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 65.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 0.6 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)