+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1703 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Intracellular part of the desmosome | |||||||||
 Map data Map data | sub-Tomogram Average | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | desmosome / cryo-electron tomography / vitreous sections | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 32.0 Å | |||||||||
 Authors Authors | Al-Amoudi A / Frangakis AS | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2011 Journal: Proc Natl Acad Sci U S A / Year: 2011Title: The three-dimensional molecular structure of the desmosomal plaque. Authors: Ashraf Al-Amoudi / Daniel Castaño-Diez / Damien P Devos / Robert B Russell / Graham T Johnson / Achilleas S Frangakis /  Abstract: The cytoplasmic surface of intercellular junctions is a complex network of molecular interactions that link the extracellular region of the desmosomal cadherins with the cytoskeletal intermediate ...The cytoplasmic surface of intercellular junctions is a complex network of molecular interactions that link the extracellular region of the desmosomal cadherins with the cytoskeletal intermediate filaments. Although 3D structures of the major plaque components are known, the overall architecture remains unknown. We used cryoelectron tomography of vitreous sections from human epidermis to record 3D images of desmosomes in vivo and in situ at molecular resolution. Our results show that the architecture of the cytoplasmic surface of the desmosome is a 2D interconnected quasiperiodic lattice, with a similar spatial organization to the extracellular side. Subtomogram averaging of the plaque region reveals two distinct layers of the desmosomal plaque: a low-density layer closer to the membrane and a high-density layer further away from the membrane. When combined with a heuristic, allowing simultaneous constrained fitting of the high-resolution structures of the major plaque proteins (desmoplakin, plakophilin, and plakoglobin), it reveals their mutual molecular interactions and explains their stoichiometry. The arrangement suggests that alternate plakoglobin-desmoplakin complexes create a template on which desmosomal cadherins cluster before they stabilize extracellularly by binding at their N-terminal tips. Plakophilins are added as a molecular reinforcement to fill the gap between the formed plaque complexes and the plasma membrane. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1703.map.gz emd_1703.map.gz | 500.4 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1703-v30.xml emd-1703-v30.xml emd-1703.xml emd-1703.xml | 10.8 KB 10.8 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-1703.png EMD-1703.png | 109.4 KB | ||
| Masks |  emd_1703_msk_1.map emd_1703_msk_1.map emd_1703_msk_2.map emd_1703_msk_2.map | 538.3 KB 538.3 KB |  Mask map Mask map | |
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1703 http://ftp.pdbj.org/pub/emdb/structures/EMD-1703 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1703 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1703 | HTTPS FTP |
-Validation report
| Summary document |  emd_1703_validation.pdf.gz emd_1703_validation.pdf.gz | 220.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1703_full_validation.pdf.gz emd_1703_full_validation.pdf.gz | 219.6 KB | Display | |
| Data in XML |  emd_1703_validation.xml.gz emd_1703_validation.xml.gz | 3.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1703 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1703 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1703 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1703 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1703.map.gz / Format: CCP4 / Size: 526.4 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1703.map.gz / Format: CCP4 / Size: 526.4 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sub-Tomogram Average | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 6 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
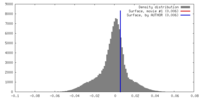
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Segmentation: Masked sub-tomogram average for residual noise removal
| Annotation | Masked sub-tomogram average for residual noise removal | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| File |  emd_1703_msk_1.map emd_1703_msk_1.map | ||||||||||||
| Projections & Slices |
| ||||||||||||
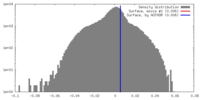
| Density Histograms |
-Segmentation: symmetrized sub-tomogram average
| Annotation | symmetrized sub-tomogram average | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| File |  emd_1703_msk_2.map emd_1703_msk_2.map | ||||||||||||
| Projections & Slices |
| ||||||||||||
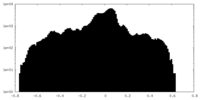
| Density Histograms |
- Sample components
Sample components
-Entire : native skin tissue
| Entire | Name: native skin tissue |
|---|---|
| Components |
|
-Supramolecule #1000: native skin tissue
| Supramolecule | Name: native skin tissue / type: sample / ID: 1000 / Number unique components: 2 |
|---|
-Supramolecule #1: desmosome
| Supramolecule | Name: desmosome / type: organelle_or_cellular_component / ID: 1 / Name.synonym: desmosome / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: NITROGEN / Instrument: OTHER |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: HOME BUILD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 32.0 Å / Resolution method: FSC 0.5 CUT-OFF |
|---|
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)