[English] 日本語
 Yorodumi
Yorodumi- EMDB-16708: HIV-1 mature capsid hexamer next to pentamer (type I) from CA-IP6... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | HIV-1 mature capsid hexamer next to pentamer (type I) from CA-IP6 CLPs bound to Nup153 peptide. | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Capsid / Hexamer / Pentamer / HIV-1 / Mature / Nup153 / VIRUS LIKE PARTICLE | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of RNA export from nucleus / nuclear pore complex assembly / Nuclear Pore Complex (NPC) Disassembly / nuclear inclusion body / nuclear pore nuclear basket / Transport of Ribonucleoproteins into the Host Nucleus / Regulation of Glucokinase by Glucokinase Regulatory Protein / Defective TPR may confer susceptibility towards thyroid papillary carcinoma (TPC) / Transport of the SLBP independent Mature mRNA / Transport of the SLBP Dependant Mature mRNA ...negative regulation of RNA export from nucleus / nuclear pore complex assembly / Nuclear Pore Complex (NPC) Disassembly / nuclear inclusion body / nuclear pore nuclear basket / Transport of Ribonucleoproteins into the Host Nucleus / Regulation of Glucokinase by Glucokinase Regulatory Protein / Defective TPR may confer susceptibility towards thyroid papillary carcinoma (TPC) / Transport of the SLBP independent Mature mRNA / Transport of the SLBP Dependant Mature mRNA / NS1 Mediated Effects on Host Pathways / SUMOylation of SUMOylation proteins / Transport of Mature mRNA Derived from an Intronless Transcript / structural constituent of nuclear pore / Rev-mediated nuclear export of HIV RNA / SUMOylation of RNA binding proteins / Nuclear import of Rev protein / RNA export from nucleus / NEP/NS2 Interacts with the Cellular Export Machinery / tRNA processing in the nucleus / Transport of Mature mRNA derived from an Intron-Containing Transcript / nucleocytoplasmic transport / Viral Messenger RNA Synthesis / nuclear localization sequence binding / SUMOylation of ubiquitinylation proteins / Vpr-mediated nuclear import of PICs / SUMOylation of DNA replication proteins / Regulation of HSF1-mediated heat shock response / mRNA transport / nuclear pore / SUMOylation of DNA damage response and repair proteins / protein-membrane adaptor activity / SUMOylation of chromatin organization proteins / nuclear periphery / HCMV Late Events / Transcriptional regulation by small RNAs / molecular condensate scaffold activity / ISG15 antiviral mechanism / HCMV Early Events / viral penetration into host nucleus / protein import into nucleus / host cell / nuclear envelope / snRNP Assembly / viral nucleocapsid / nuclear membrane / host cell cytoplasm / amyloid fibril formation / symbiont entry into host cell / viral translational frameshifting / host cell nucleus / nucleolus / SARS-CoV-2 activates/modulates innate and adaptive immune responses / structural molecule activity / virion membrane / DNA binding / RNA binding / zinc ion binding / nucleoplasm / ATP binding / identical protein binding / membrane / metal ion binding / cytosol / cytoplasm Similarity search - Function | ||||||||||||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.12 Å | ||||||||||||||||||
 Authors Authors | Stacey JCV / Briggs JAG | ||||||||||||||||||
| Funding support |  Germany, Germany,  United States, United States,  United Kingdom, 5 items United Kingdom, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Two structural switches in HIV-1 capsid regulate capsid curvature and host factor binding. Authors: James C V Stacey / Aaron Tan / John M Lu / Leo C James / Robert A Dick / John A G Briggs /    Abstract: The mature HIV-1 capsid protects the viral genome and interacts with host proteins to travel from the cell periphery into the nucleus. To achieve this, the capsid protein, CA, constructs conical ...The mature HIV-1 capsid protects the viral genome and interacts with host proteins to travel from the cell periphery into the nucleus. To achieve this, the capsid protein, CA, constructs conical capsids from a lattice of hexamers and pentamers, and engages in and then relinquishes multiple interactions with cellular proteins in an orchestrated fashion. Cellular host factors including Nup153, CPSF6, and Sec24C engage the same pocket within CA hexamers. How CA assembles pentamers and hexamers of different curvatures, how CA oligomerization states or curvature might modulate host-protein interactions, and how binding of multiple cofactors to a single site is coordinated, all remain to be elucidated. Here, using single-particle cryoEM, we have determined the structure of the mature HIV-1 CA pentamer and hexamer from conical CA-IP polyhedra to ~3 Å resolution. We also determined structures of hexamers in the context of multiple lattice curvatures and number of pentamer contacts. Comparison of these structures, bound or not to host protein peptides, revealed two structural switches within HIV-1 CA that modulate peptide binding according to CA lattice curvature and whether CA is hexameric or pentameric. These observations suggest that the conical HIV-1 capsid has different host-protein binding properties at different positions on its surface, which may facilitate cell entry and represent an evolutionary advantage of conical morphology. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16708.map.gz emd_16708.map.gz | 203.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16708-v30.xml emd-16708-v30.xml emd-16708.xml emd-16708.xml | 21.6 KB 21.6 KB | Display Display |  EMDB header EMDB header |
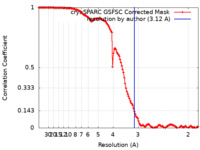
| FSC (resolution estimation) |  emd_16708_fsc.xml emd_16708_fsc.xml | 14.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_16708.png emd_16708.png | 94.4 KB | ||
| Masks |  emd_16708_msk_1.map emd_16708_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16708.cif.gz emd-16708.cif.gz | 6.7 KB | ||
| Others |  emd_16708_half_map_1.map.gz emd_16708_half_map_1.map.gz emd_16708_half_map_2.map.gz emd_16708_half_map_2.map.gz | 200.2 MB 200.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16708 http://ftp.pdbj.org/pub/emdb/structures/EMD-16708 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16708 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16708 | HTTPS FTP |
-Validation report
| Summary document |  emd_16708_validation.pdf.gz emd_16708_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16708_full_validation.pdf.gz emd_16708_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_16708_validation.xml.gz emd_16708_validation.xml.gz | 21.3 KB | Display | |
| Data in CIF |  emd_16708_validation.cif.gz emd_16708_validation.cif.gz | 27.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16708 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16708 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16708 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16708 | HTTPS FTP |
-Related structure data
| Related structure data |  8cl0MC  8ckvC  8ckwC  8ckxC  8ckyC  8ckzC  8cl1C  8cl2C  8cl3C  8cl4C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16708.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16708.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.93 Å | ||||||||||||||||||||||||||||||||||||
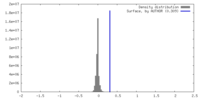
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16708_msk_1.map emd_16708_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
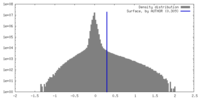
| Density Histograms |
-Half map: #2
| File | emd_16708_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
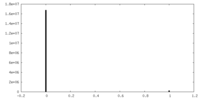
| Density Histograms |
-Half map: #1
| File | emd_16708_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
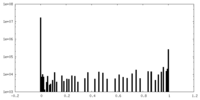
| Density Histograms |
- Sample components
Sample components
-Entire : HIV-1 Mature Capsid
| Entire | Name: HIV-1 Mature Capsid |
|---|---|
| Components |
|
-Supramolecule #1: HIV-1 Mature Capsid
| Supramolecule | Name: HIV-1 Mature Capsid / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Pentamer adjacent Hexamer (type I) bound to FG repeat containing Nup153 peptide. |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 27 KDa |
-Macromolecule #1: Gag polyprotein
| Macromolecule | Name: Gag polyprotein / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 25.761623 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPIVQNLQGQ MVHQAISPRT LNAWVKVVEE KAFSPEVIPM FSALSEGATP QDLNTMLNTV GGHQAAMQML KETINEEAAE WDRLHPVHA GPIAPGQMRE PRGSDIAGTT STLQEQIGWM THNPPIPVGE IYKRWIILGL NKIVRMYSPT SILDIRQGPK E PFRDYVDR ...String: MPIVQNLQGQ MVHQAISPRT LNAWVKVVEE KAFSPEVIPM FSALSEGATP QDLNTMLNTV GGHQAAMQML KETINEEAAE WDRLHPVHA GPIAPGQMRE PRGSDIAGTT STLQEQIGWM THNPPIPVGE IYKRWIILGL NKIVRMYSPT SILDIRQGPK E PFRDYVDR FYKTLRAEQA SQEVKNWMTE TLLVQNANPD CKTILKALGP GATLEEMMTA CQGVGGPGHK ARVL UniProtKB: Gag polyprotein |
-Macromolecule #2: Nuclear pore complex protein Nup153
| Macromolecule | Name: Nuclear pore complex protein Nup153 / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.687721 KDa |
| Sequence | String: TNNSPSGVFT FGANSST UniProtKB: Nuclear pore complex protein Nup153 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.1 Component:
| ||||||||||||||||||
| Grid | Model: C-flat-2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 2 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||||||||
| Details | VLP assembly, bound to Nup153 peptide. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Cross-correlation | ||||||
| Output model |  PDB-8cl0: |
 Movie
Movie Controller
Controller































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)