[English] 日本語
 Yorodumi
Yorodumi- EMDB-16309: In situ outer dynein arm from Chlamydomonas reinhardtii in an int... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
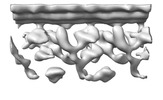
| Title | In situ outer dynein arm from Chlamydomonas reinhardtii in an intermediate state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | axoneme / outer dynein arm / power stroke / dynein / MOTOR PROTEIN | |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 39.8 Å | |||||||||
 Authors Authors | Zimmermann NEL / Noga A / Obbineni JM / Ishikawa T | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2023 Journal: EMBO J / Year: 2023Title: ATP-induced conformational change of axonemal outer dynein arms revealed by cryo-electron tomography. Authors: Noemi Zimmermann / Akira Noga / Jagan Mohan Obbineni / Takashi Ishikawa /   Abstract: Axonemal outer dynein arm (ODA) motors generate force for ciliary beating. We analyzed three states of the ODA during the power stroke cycle using in situ cryo-electron tomography, subtomogram ...Axonemal outer dynein arm (ODA) motors generate force for ciliary beating. We analyzed three states of the ODA during the power stroke cycle using in situ cryo-electron tomography, subtomogram averaging, and classification. These states of force generation depict the prepower stroke, postpower stroke, and intermediate state conformations. Comparison of these conformations to published in vitro atomic structures of cytoplasmic dynein, ODA, and the Shulin-ODA complex revealed differences in the orientation and position of the dynein head. Our analysis shows that in the absence of ATP, all dynein linkers interact with the AAA3/AAA4 domains, indicating that interactions with the adjacent microtubule doublet B-tubule direct dynein orientation. For the prepower stroke conformation, there were changes in the tail that is anchored on the A-tubule. We built models starting with available high-resolution structures to generate a best-fitting model structure for the in situ pre- and postpower stroke ODA conformations, thereby showing that ODA in a complex with Shulin adopts a similar conformation as the active prepower stroke ODA in the axoneme. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16309.map.gz emd_16309.map.gz | 1.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16309-v30.xml emd-16309-v30.xml emd-16309.xml emd-16309.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16309.png emd_16309.png | 49.6 KB | ||
| Others |  emd_16309_half_map_1.map.gz emd_16309_half_map_1.map.gz emd_16309_half_map_2.map.gz emd_16309_half_map_2.map.gz | 1.4 MB 1.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16309 http://ftp.pdbj.org/pub/emdb/structures/EMD-16309 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16309 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16309 | HTTPS FTP |
-Validation report
| Summary document |  emd_16309_validation.pdf.gz emd_16309_validation.pdf.gz | 841.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16309_full_validation.pdf.gz emd_16309_full_validation.pdf.gz | 840.7 KB | Display | |
| Data in XML |  emd_16309_validation.xml.gz emd_16309_validation.xml.gz | 6.9 KB | Display | |
| Data in CIF |  emd_16309_validation.cif.gz emd_16309_validation.cif.gz | 7.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16309 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16309 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16309 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16309 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16309.map.gz / Format: CCP4 / Size: 1.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16309.map.gz / Format: CCP4 / Size: 1.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 8.5 Å | ||||||||||||||||||||||||||||||||||||
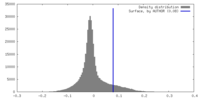
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_16309_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
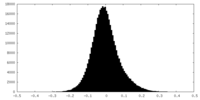
| Density Histograms |
-Half map: #2
| File | emd_16309_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
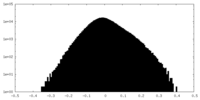
| Density Histograms |
- Sample components
Sample components
-Entire : In situ outer dynein arm
| Entire | Name: In situ outer dynein arm |
|---|---|
| Components |
|
-Supramolecule #1: In situ outer dynein arm
| Supramolecule | Name: In situ outer dynein arm / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1-#21 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 4.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: BACK PROJECTION / Resolution.type: BY AUTHOR / Resolution: 39.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number subtomograms used: 265 |
|---|---|
| Extraction | Number tomograms: 12 / Number images used: 3553 |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)