+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the CODV-IL13-RefAb triple complex | |||||||||
 Map data Map data | Pre-sharpened, unmasked map of CODV-IL13-RefAb after non-uniform refinement | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cytosolic / bispecific / Fab / therapeutical / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of complement-dependent cytotoxicity / cytokine receptor binding / negative regulation of endothelial cell apoptotic process / immune response / extracellular region Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
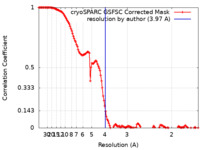
| Method | single particle reconstruction / cryo EM / Resolution: 3.97 Å | |||||||||
 Authors Authors | Fernandez-Martinez D / Kandiah E | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2023 Journal: Sci Rep / Year: 2023Title: Structural insights into the bi-specific cross-over dual variable antibody architecture by cryo-EM. Authors: David Fernandez-Martinez / Mark D Tully / Gordon Leonard / Magali Mathieu / Eaazhisai Kandiah /  Abstract: Multi-specific antibodies (msAbs) are being developed as next generation antibody-based therapeutics. Knowledge of the three-dimensional structures, in the full antibody context, of their fragment ...Multi-specific antibodies (msAbs) are being developed as next generation antibody-based therapeutics. Knowledge of the three-dimensional structures, in the full antibody context, of their fragment antigen-binding (Fab) moieties with or without bound antigens is key to elucidating their therapeutic efficiency and stability. However, the flexibility of msAbs, a feature essential for their multi specificity, has hindered efforts in this direction. Cross-Over Dual Variable immunoglobulin (CODV) is a promising bispecific antibody format, designed to simultaneously target the interleukins IL4 and IL13. In this work we present the biophysical and structural characterisation of a CODV:IL13 complex in the full antibody context, using cryo-electron microscopy at an overall resolution of 4.2 Å. Unlike the 1:2 stoichiometry previously observed for CODV:IL4, CODV:IL13 shows a 1:1 stoichiometry. As well as providing details of the IL13-CODV binding interface, including the residues involved in the epitope-paratope region, the structure of CODV:IL13 also validates the use of labelling antibody as a new strategy for the single particle cryo-EM study of msAbs in complex with one, or more, antigens. This strategy reduced the inherent flexibility of the IL13 binding domain of CODV without inducing either structural changes at the epitope level or steric hindrance between the IL4 and IL13 binding regions of CODV. The work presented here thus also contributes to the development of methodology for the structural study of msAbs, a promising platform for cancer immunotherapy. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16113.map.gz emd_16113.map.gz | 120 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16113-v30.xml emd-16113-v30.xml emd-16113.xml emd-16113.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16113_fsc.xml emd_16113_fsc.xml | 15 KB | Display |  FSC data file FSC data file |
| Images |  emd_16113.png emd_16113.png | 46 KB | ||
| Masks |  emd_16113_msk_1.map emd_16113_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Others |  emd_16113_additional_1.map.gz emd_16113_additional_1.map.gz emd_16113_half_map_1.map.gz emd_16113_half_map_1.map.gz emd_16113_half_map_2.map.gz emd_16113_half_map_2.map.gz | 217.1 MB 226.3 MB 226.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16113 http://ftp.pdbj.org/pub/emdb/structures/EMD-16113 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16113 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16113 | HTTPS FTP |
-Validation report
| Summary document |  emd_16113_validation.pdf.gz emd_16113_validation.pdf.gz | 791.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16113_full_validation.pdf.gz emd_16113_full_validation.pdf.gz | 791.4 KB | Display | |
| Data in XML |  emd_16113_validation.xml.gz emd_16113_validation.xml.gz | 21.7 KB | Display | |
| Data in CIF |  emd_16113_validation.cif.gz emd_16113_validation.cif.gz | 27.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16113 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16113 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16113 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16113 | HTTPS FTP |
-Related structure data
| Related structure data |  8blqMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16113.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16113.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Pre-sharpened, unmasked map of CODV-IL13-RefAb after non-uniform refinement | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.827 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16113_msk_1.map emd_16113_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
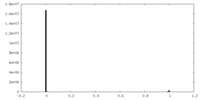
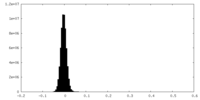
| Density Histograms |
-Additional map: Final sharpened and masked map of CODV-IL13-RefAb
| File | emd_16113_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final sharpened and masked map of CODV-IL13-RefAb | ||||||||||||
| Projections & Slices |
| ||||||||||||
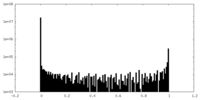
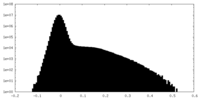
| Density Histograms |
-Half map: Half map B of the pre-sharpened, unmasked map...
| File | emd_16113_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B of the pre-sharpened, unmasked map of CODV-IL13-RefAb during non-uniform refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
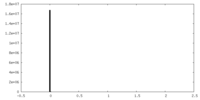
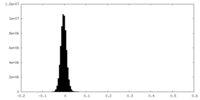
| Density Histograms |
-Half map: Half map A of the pre-sharpened, unmasked map...
| File | emd_16113_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A of the pre-sharpened, unmasked map of CODV-IL13-RefAb during non-uniform refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
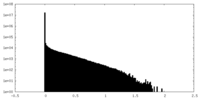
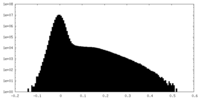
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of CODV-Fab with IL13 and RefAb
| Entire | Name: Ternary complex of CODV-Fab with IL13 and RefAb |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of CODV-Fab with IL13 and RefAb
| Supramolecule | Name: Ternary complex of CODV-Fab with IL13 and RefAb / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: CODV-Fab, heavy chain
| Macromolecule | Name: CODV-Fab, heavy chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 36.907098 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VQLQQSGPEL VKPGASVKIS CKASGYSFTS YWIHWIKQRP GQGLEWIGMI DPSDGETRLN QRFQGRATLT VDESTSTAYM QLRSPTSED SAVYYCTRLK EYGNYDSFYF DVWGAGTLVT VSSGEVQLKE SGPGLVAPGG SLSITCTVSG FSLTDSSINW V RQPPGKGL ...String: VQLQQSGPEL VKPGASVKIS CKASGYSFTS YWIHWIKQRP GQGLEWIGMI DPSDGETRLN QRFQGRATLT VDESTSTAYM QLRSPTSED SAVYYCTRLK EYGNYDSFYF DVWGAGTLVT VSSGEVQLKE SGPGLVAPGG SLSITCTVSG FSLTDSSINW V RQPPGKGL EWLGMIWGDG RIDYADALKS RLSISKDSSK SQVFLEMTSL RTDDTATYYC ARDGYFPYAM DFWGQGTSVT VS SGGASTK GPSVFPLAPS SKSTSGGTAA LGCLVKDYFP EPVTVSWNSG ALTSGVHTFP AVLQSSGLYS LSSVVTVPSS SLG TQTYIC NVNHKPSNTK VDKKVEP |
-Macromolecule #2: CODV-Fab, light chain
| Macromolecule | Name: CODV-Fab, light chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 35.726516 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVLTQSPAS LAVSLGQRAT ISCRASESVD SYGQSYMHWY QQKAGQPPKL LIYLASNLES GVPARFSGSG SRTDFTLTID PVQAEDAAT YYCQQNAEDS RTFGGGTKLE IKGGGGGGGD IQMTQSPASL SVSVGDTITL TCHASQNIDV WLSWFQQKPG N IPKLLIYK ...String: DIVLTQSPAS LAVSLGQRAT ISCRASESVD SYGQSYMHWY QQKAGQPPKL LIYLASNLES GVPARFSGSG SRTDFTLTID PVQAEDAAT YYCQQNAEDS RTFGGGTKLE IKGGGGGGGD IQMTQSPASL SVSVGDTITL TCHASQNIDV WLSWFQQKPG N IPKLLIYK ASNLHTGVPS RFSGSGSGTG FTLTISSLQP EDIATYYCQQ AHSYPFTFGG GTKLEIKGGG GGRTVAAPSV FI FPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL QSGNSQESVT EQDSKDSTYS LSSTLTLSKA DYEKHKVYAC EVT HQGLSS PVTKSFNR |
-Macromolecule #3: RefAb, light chain
| Macromolecule | Name: RefAb, light chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.51002 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: YVLTQPPSVS VAPGKTARIT CGGNIIGSKL VHWYQQKPGQ APVLVIYDDG DRPSGIPERF SGSNSGNTAT LTISRVEAGD EADYYCQVW DTGSDPVVFG GGTKLTVLGQ PKAAPSVTLF PPSSEELQAN KATLVCLISD FYPGAVTVAW KADSSPVKAG V ETTTPSKQ ...String: YVLTQPPSVS VAPGKTARIT CGGNIIGSKL VHWYQQKPGQ APVLVIYDDG DRPSGIPERF SGSNSGNTAT LTISRVEAGD EADYYCQVW DTGSDPVVFG GGTKLTVLGQ PKAAPSVTLF PPSSEELQAN KATLVCLISD FYPGAVTVAW KADSSPVKAG V ETTTPSKQ SNNKYAASSY LSLTPEQWKS HRSYSCQVTH EGSTVEKTVA PTEC |
-Macromolecule #4: Interleukin-13, human
| Macromolecule | Name: Interleukin-13, human / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.077022 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: PPSTALRELI EELVNITQNQ KAPLCNGSMV WSINLTAGMY CAALESLINV SGCSAIEKTQ RMLSGFCPHK VSAGQFSSLH VRDTKIEVA QFVKDLLLHL KKLFREGQFN |
-Macromolecule #5: RefAb, heavy chain
| Macromolecule | Name: RefAb, heavy chain / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.751457 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VQLVQSGAEV KKPGASVKVS CKASGYTFTN YGLSWVRQAP GQGLEWMGWI SANNGDTNYG QEFQGRVTMT TDTSTSTAYM ELRSLRSDD TAVYYCARDS SSSWARWFFD LWGRGTLVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL VKDYFPEPVT V SWNSGALT ...String: VQLVQSGAEV KKPGASVKVS CKASGYTFTN YGLSWVRQAP GQGLEWMGWI SANNGDTNYG QEFQGRVTMT TDTSTSTAYM ELRSLRSDD TAVYYCARDS SSSWARWFFD LWGRGTLVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL VKDYFPEPVT V SWNSGALT SGVHTFPAVL QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKK VEPK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 46.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8blq: |
 Movie
Movie Controller
Controller







 Z
Z Y
Y X
X