+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1600 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Motor mechanism for protein threading through Hsp104 | |||||||||
 Map data Map data | Yeast Hsp104 N728A 3D density map. Hexamer formed in the presence of ATP. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Clp/Hsp100 / AAA+ / protein remodeling / disaggregation / molecular machines / heat shock protein / Hsp104 / ClpB / yeast prions | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 11.5 Å | |||||||||
 Authors Authors | Wendler P / Shorter J / Snead D / Plisson C / Clare DK / Lindquist S / Saibil H | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2009 Journal: Mol Cell / Year: 2009Title: Motor mechanism for protein threading through Hsp104. Authors: Petra Wendler / James Shorter / David Snead / Celia Plisson / Daniel K Clare / Susan Lindquist / Helen R Saibil /  Abstract: The protein-remodeling machine Hsp104 dissolves amorphous aggregates as well as ordered amyloid assemblies such as yeast prions. Force generation originates from a tandem AAA+ (ATPases associated ...The protein-remodeling machine Hsp104 dissolves amorphous aggregates as well as ordered amyloid assemblies such as yeast prions. Force generation originates from a tandem AAA+ (ATPases associated with various cellular activities) cassette, but the mechanism and allostery of this action remain to be established. Our cryoelectron microscopy maps of Hsp104 hexamers reveal substantial domain movements upon ATP binding and hydrolysis in the first nucleotide-binding domain (NBD1). Fitting atomic models of Hsp104 domains to the EM density maps plus supporting biochemical measurements show how the domain movements displace sites bearing the substrate-binding tyrosine loops. This provides the structural basis for N- to C-terminal substrate threading through the central cavity, enabling a clockwise handover of substrate in the NBD1 ring and coordinated substrate binding between NBD1 and NBD2. Asymmetric reconstructions of Hsp104 in the presence of ATPgammaS or ATP support sequential rather than concerted ATP hydrolysis in the NBD1 ring. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1600.map.gz emd_1600.map.gz | 311.5 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1600-v30.xml emd-1600-v30.xml emd-1600.xml emd-1600.xml | 9.6 KB 9.6 KB | Display Display |  EMDB header EMDB header |
| Images |  1600.png 1600.png | 338.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1600 http://ftp.pdbj.org/pub/emdb/structures/EMD-1600 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1600 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1600 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1600.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1600.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Yeast Hsp104 N728A 3D density map. Hexamer formed in the presence of ATP. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
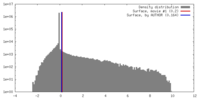
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : S. cerevisiae Hsp104 N728A ATP
| Entire | Name: S. cerevisiae Hsp104 N728A ATP |
|---|---|
| Components |
|
-Supramolecule #1000: S. cerevisiae Hsp104 N728A ATP
| Supramolecule | Name: S. cerevisiae Hsp104 N728A ATP / type: sample / ID: 1000 / Oligomeric state: hexamer / Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 612 KDa |
-Macromolecule #1: Hsp104 N728A
| Macromolecule | Name: Hsp104 N728A / type: protein_or_peptide / ID: 1 / Name.synonym: Hsp104 N728A / Details: Hexamers were formed in the presence of 5mM ADP / Number of copies: 6 / Oligomeric state: hexamer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 102 KDa |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 20 mM HEPES pH 7.5, 20 mM NaCl, 10 mM MgCl2, 1 mM DTT, 2-5mM ATP |
| Grid | Details: 300 mesh copper grid- holey carbon film |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: home made Method: The grids were blotted for 2-3 sec and immediately plunged into liquid ethane |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Min: 77 K / Max: 85 K / Average: 77 K |
| Alignment procedure | Legacy - Astigmatism: corrected for at specimen level |
| Date | Jan 28, 2004 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 21 / Average electron dose: 15 e/Å2 / Od range: 1 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 50000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: single tilt cryo / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: phase flipping, each particle |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C6 (6 fold cyclic) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 11.5 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: IMAGIC SPIDER MRC / Details: 6-fold symmetrised reconstruction / Number images used: 4046 |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)