[English] 日本語
 Yorodumi
Yorodumi- EMDB-1589: Bacterial dynamin-like protein undergoes radical conformational c... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1589 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bacterial dynamin-like protein undergoes radical conformational changes that promote membrane curvature | |||||||||
 Map data Map data | Bacterial dynamin BDLP lipid tube reconstruction, single-particle cryo EM helical reconstruction | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | dynamin / membrane / GTPase | |||||||||
| Function / homology |  Function and homology information Function and homology informationdynamin GTPase / mitochondrial fusion / GTPase activity / lipid binding / GTP binding / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Nostoc punctiforme (bacteria) Nostoc punctiforme (bacteria) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 9.0 Å | |||||||||
 Authors Authors | Low HH / Sachse C / Amos LA / Lowe J | |||||||||
 Citation Citation |  Journal: Cell / Year: 2009 Journal: Cell / Year: 2009Title: Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Authors: Harry H Low / Carsten Sachse / Linda A Amos / Jan Löwe /  Abstract: Proteins of the dynamin superfamily mediate membrane fission, fusion, and restructuring events by polymerizing upon lipid bilayers and forcing regions of high curvature. In this work, we show the ...Proteins of the dynamin superfamily mediate membrane fission, fusion, and restructuring events by polymerizing upon lipid bilayers and forcing regions of high curvature. In this work, we show the electron cryomicroscopy reconstruction of a bacterial dynamin-like protein (BDLP) helical filament decorating a lipid tube at approximately 11 A resolution. We fitted the BDLP crystal structure and produced a molecular model for the entire filament. The BDLP GTPase domain dimerizes and forms the tube surface, the GTPase effector domain (GED) mediates self-assembly, and the paddle region contacts the lipids and promotes curvature. Association of BDLP with GMPPNP and lipid induces radical, large-scale conformational changes affecting polymerization. Nucleotide hydrolysis seems therefore to be coupled to polymer disassembly and dissociation from lipid, rather than membrane restructuring. Observed structural similarities with rat dynamin 1 suggest that our results have broad implication for other dynamin family members. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1589.map.gz emd_1589.map.gz | 11.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1589-v30.xml emd-1589-v30.xml emd-1589.xml emd-1589.xml | 8.6 KB 8.6 KB | Display Display |  EMDB header EMDB header |
| Images |  1589.png 1589.png | 307.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1589 http://ftp.pdbj.org/pub/emdb/structures/EMD-1589 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1589 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1589 | HTTPS FTP |
-Validation report
| Summary document |  emd_1589_validation.pdf.gz emd_1589_validation.pdf.gz | 219.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1589_full_validation.pdf.gz emd_1589_full_validation.pdf.gz | 218.3 KB | Display | |
| Data in XML |  emd_1589_validation.xml.gz emd_1589_validation.xml.gz | 4.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1589 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1589 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1589 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1589 | HTTPS FTP |
-Related structure data
| Related structure data |  2w6dMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1589.map.gz / Format: CCP4 / Size: 32.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1589.map.gz / Format: CCP4 / Size: 32.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Bacterial dynamin BDLP lipid tube reconstruction, single-particle cryo EM helical reconstruction | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.03398 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
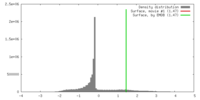
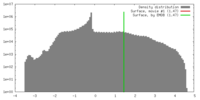
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Bacterial dynamin-like protein BDLP
| Entire | Name: Bacterial dynamin-like protein BDLP |
|---|---|
| Components |
|
-Supramolecule #1000: Bacterial dynamin-like protein BDLP
| Supramolecule | Name: Bacterial dynamin-like protein BDLP / type: sample / ID: 1000 Oligomeric state: BDLP dimer is the asymmetric unit of the tubes Number unique components: 1 |
|---|
-Macromolecule #1: Bacterial dynamin-like protein BDLP
| Macromolecule | Name: Bacterial dynamin-like protein BDLP / type: protein_or_peptide / ID: 1 / Name.synonym: BDLP / Oligomeric state: dimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Nostoc punctiforme (bacteria) Nostoc punctiforme (bacteria) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE / Instrument: OTHER |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Category: FILM / Film or detector model: GENERIC FILM / Digitization - Scanner: OTHER |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 59000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: OTHER |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 3.1926 Å Applied symmetry - Helical parameters - Δ&Phi: 63.815 ° Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 9.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER |
|---|---|
| CTF correction | Details: CTF2, CTFTILT |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)