[English] 日本語
 Yorodumi
Yorodumi- EMDB-15801: cryo-EM structure of carboxysomal mini-shell: icosahedral assembl... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of carboxysomal mini-shell: icosahedral assembly from CsoS4A/1A and CsoS2 co-expression (T = 9) | |||||||||
 Map data Map data | main map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | carboxysome / shell / icosahedral symmetry / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationstructural constituent of carboxysome shell / carboxysome / carbon fixation / viral translational frameshifting Similarity search - Function | |||||||||
| Biological species |  Halothiobacillus neapolitanus (bacteria) Halothiobacillus neapolitanus (bacteria) | |||||||||
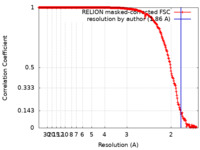
| Method | single particle reconstruction / cryo EM / Resolution: 1.86 Å | |||||||||
 Authors Authors | Ni T / Jiang Q / Liu LN / Zhang P | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Intrinsically disordered CsoS2 acts as a general molecular thread for α-carboxysome shell assembly. Authors: Tao Ni / Qiuyao Jiang / Pei Cing Ng / Juan Shen / Hao Dou / Yanan Zhu / Julika Radecke / Gregory F Dykes / Fang Huang / Lu-Ning Liu / Peijun Zhang /   Abstract: Carboxysomes are a paradigm of self-assembling proteinaceous organelles found in nature, offering compartmentalisation of enzymes and pathways to enhance carbon fixation. In α-carboxysomes, the ...Carboxysomes are a paradigm of self-assembling proteinaceous organelles found in nature, offering compartmentalisation of enzymes and pathways to enhance carbon fixation. In α-carboxysomes, the disordered linker protein CsoS2 plays an essential role in carboxysome assembly and Rubisco encapsulation. Its mechanism of action, however, is not fully understood. Here we synthetically engineer α-carboxysome shells using minimal shell components and determine cryoEM structures of these to decipher the principle of shell assembly and encapsulation. The structures reveal that the intrinsically disordered CsoS2 C-terminus is well-structured and acts as a universal "molecular thread" stitching through multiple shell protein interfaces. We further uncover in CsoS2 a highly conserved repetitive key interaction motif, [IV]TG, which is critical to the shell assembly and architecture. Our study provides a general mechanism for the CsoS2-governed carboxysome shell assembly and cargo encapsulation and further advances synthetic engineering of carboxysomes for diverse biotechnological applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15801.map.gz emd_15801.map.gz | 1.3 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15801-v30.xml emd-15801-v30.xml emd-15801.xml emd-15801.xml | 18.9 KB 18.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15801_fsc.xml emd_15801_fsc.xml | 25.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_15801.png emd_15801.png | 227.8 KB | ||
| Masks |  emd_15801_msk_1.map emd_15801_msk_1.map | 1.4 GB |  Mask map Mask map | |
| Filedesc metadata |  emd-15801.cif.gz emd-15801.cif.gz | 6.4 KB | ||
| Others |  emd_15801_half_map_1.map.gz emd_15801_half_map_1.map.gz emd_15801_half_map_2.map.gz emd_15801_half_map_2.map.gz | 1.1 GB 1.1 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15801 http://ftp.pdbj.org/pub/emdb/structures/EMD-15801 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15801 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15801 | HTTPS FTP |
-Validation report
| Summary document |  emd_15801_validation.pdf.gz emd_15801_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15801_full_validation.pdf.gz emd_15801_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_15801_validation.xml.gz emd_15801_validation.xml.gz | 32.3 KB | Display | |
| Data in CIF |  emd_15801_validation.cif.gz emd_15801_validation.cif.gz | 43.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15801 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15801 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15801 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15801 | HTTPS FTP |
-Related structure data
| Related structure data |  8b12MC  8b0yC  8b11C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15801.map.gz / Format: CCP4 / Size: 1.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15801.map.gz / Format: CCP4 / Size: 1.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.831 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15801_msk_1.map emd_15801_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_15801_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_15801_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : mini-shell assembly from Halothiobacillus neapolitanus with CsoS4...
| Entire | Name: mini-shell assembly from Halothiobacillus neapolitanus with CsoS4A/1A and CsoS2 co-expression (T= 9) |
|---|---|
| Components |
|
-Supramolecule #1: mini-shell assembly from Halothiobacillus neapolitanus with CsoS4...
| Supramolecule | Name: mini-shell assembly from Halothiobacillus neapolitanus with CsoS4A/1A and CsoS2 co-expression (T= 9) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Halothiobacillus neapolitanus (bacteria) Halothiobacillus neapolitanus (bacteria) |
-Macromolecule #1: Major carboxysome shell protein CsoS1A
| Macromolecule | Name: Major carboxysome shell protein CsoS1A / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Halothiobacillus neapolitanus (bacteria) / Strain: ATCC 23641 / c2 Halothiobacillus neapolitanus (bacteria) / Strain: ATCC 23641 / c2 |
| Molecular weight | Theoretical: 9.973478 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MADVTGIALG MIETRGLVPA IEAADAMTKA AEVRLVGRQF VGGGYVTVLV RGETGAVNAA VRAGADACER VGDGLVAAHI IARVHSEVE NILPKAPQA UniProtKB: Major carboxysome shell protein CsoS1A |
-Macromolecule #2: Carboxysome shell vertex protein CsoS4A
| Macromolecule | Name: Carboxysome shell vertex protein CsoS4A / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Halothiobacillus neapolitanus (bacteria) / Strain: ATCC 23641 / c2 Halothiobacillus neapolitanus (bacteria) / Strain: ATCC 23641 / c2 |
| Molecular weight | Theoretical: 8.900287 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKIMQVEKTL VSTNRIADMG HKPLLVVWEK PGAPRQVAVD AIGCIPGDWV LCVGSSAARE AAGSKSYPSD LTIIGIIDQW NGE UniProtKB: Carboxysome shell vertex protein CsoS4A |
-Macromolecule #3: Carboxysome assembly protein CsoS2B
| Macromolecule | Name: Carboxysome assembly protein CsoS2B / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Halothiobacillus neapolitanus (bacteria) / Strain: ATCC 23641 / c2 Halothiobacillus neapolitanus (bacteria) / Strain: ATCC 23641 / c2 |
| Molecular weight | Theoretical: 91.502031 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNPADLSGLS GKELARARRA ALSKQGKAAV SNKTASVNRS TKQAASSINT NQVRSSVNEV PTDYQMADQL CSTIDHADFG TESNRVRDL CRQRREALST IGKKAVKTNG KPSGRVRPQQ SVVHNDAMIE NAGDTNQSSS TSLNNELSEI CSIADDMPER F GSQAKTVR ...String: MNPADLSGLS GKELARARRA ALSKQGKAAV SNKTASVNRS TKQAASSINT NQVRSSVNEV PTDYQMADQL CSTIDHADFG TESNRVRDL CRQRREALST IGKKAVKTNG KPSGRVRPQQ SVVHNDAMIE NAGDTNQSSS TSLNNELSEI CSIADDMPER F GSQAKTVR DICRARRQAL SERGTRAVPP KPQSQGGPGR NGYQIDGYLD TALHGRDAAK RHREMLCQYG RGTAPSCKPT GR VKNSVQS GNAAPKKVET GHTLSGGSVT GTQVDRKSHV TGNEPGTCRA VTGTEYVGTE QFTSFCNTSP KPNATKVNVT TTA RGRPVS GTEVSRTEKV TGNESGVCRN VTGTEYMSNE AHFSLCGTAA KPSQADKVMF GATARTHQVV SGSDEFRPSS VTGN ESGAK RTITGSQYAD EGLARLTING APAKVARTHT FAGSDVTGTE IGRSTRVTGD ESGSCRSISG TEYLSNEQFQ SFCDT KPQR SPFKVGQDRT NKGQSVTGNL VDRSELVTGN EPGSCSRVTG SQYGQSKICG GGVGKVRSMR TLRGTSVSGQ QLDHAP KMS GDERGGCMPV TGNEYYGREH FEPFCTSTPE PEAQSTEQSL TCEGQIISGT SVDASDLVTG NEIGEQQLIS GDAYVGA QQ TGCLPTSPRF NQTGNVQSMG FKNTNQPEQN FAPGEVMPTD FSIQTPARSA QNRITGNDIA PSGRITGPGM LATGLITG T PEFRHAAREL VGSPQPMAMA MANRNKAAQA PVVQPEVVAT QEKPELVCAP RSDQMDRVSG EGKERCHITG DDWSVNKHI TGTAGQWASG RNPSMRGNAR VVETSAFANR NVPKPEKPGS KITGSSGNDT QGSLITYSGG ARG UniProtKB: Carboxysome assembly protein CsoS2B |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 485 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 44.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)