[English] 日本語
 Yorodumi
Yorodumi- EMDB-1580: CryoEM and 3D image reconstruction of Chilo Iridescent virus at 1... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1580 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM and 3D image reconstruction of Chilo Iridescent virus at 13 angstrom resolution | |||||||||
 Map data Map data | Chilo Iridescent virus at 13 angstrom resolution | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | large DNA virus / cryo-electron microscopy / 3D image reconstruction / enveloped virus / minor capsid proteins | |||||||||
| Biological species |  Invertebrate iridescent virus 6 Invertebrate iridescent virus 6 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 13.0 Å | |||||||||
 Authors Authors | Yan X / Yu Z / Zhang P / Battisti AJ / Holdaway HA / Chipman PR / Bajaj C / Bergoin M / Rossmann MG / Baker TS | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2009 Journal: J Mol Biol / Year: 2009Title: The capsid proteins of a large, icosahedral dsDNA virus. Authors: Xiaodong Yan / Zeyun Yu / Ping Zhang / Anthony J Battisti / Heather A Holdaway / Paul R Chipman / Chandrajit Bajaj / Max Bergoin / Michael G Rossmann / Timothy S Baker /  Abstract: Chilo iridescent virus (CIV) is a large (approximately 1850 A diameter) insect virus with an icosahedral, T=147 capsid, a double-stranded DNA (dsDNA) genome, and an internal lipid membrane. The ...Chilo iridescent virus (CIV) is a large (approximately 1850 A diameter) insect virus with an icosahedral, T=147 capsid, a double-stranded DNA (dsDNA) genome, and an internal lipid membrane. The structure of CIV was determined to 13 A resolution by means of cryoelectron microscopy (cryoEM) and three-dimensional image reconstruction. A homology model of P50, the CIV major capsid protein (MCP), was built based on its amino acid sequence and the structure of the homologous Paramecium bursaria chlorella virus 1 Vp54 MCP. This model was fitted into the cryoEM density for each of the 25 trimeric CIV capsomers per icosahedral asymmetric unit. A difference map, in which the fitted CIV MCP capsomers were subtracted from the CIV cryoEM reconstruction, showed that there are at least three different types of minor capsid proteins associated with the capsomers outside the lipid membrane. "Finger" proteins are situated at many, but not all, of the spaces between three adjacent capsomers within each trisymmetron, and "zip" proteins are situated between sets of three adjacent capsomers at the boundary between neighboring trisymmetrons and pentasymmetrons. Based on the results of segmentation and density correlations, there are at least eight finger proteins and three dimeric and two monomeric zip proteins in one asymmetric unit of the CIV capsid. These minor proteins appear to stabilize the virus by acting as intercapsomer cross-links. One transmembrane "anchor" protein per icosahedral asymmetric unit, which extends from beneath one of the capsomers in the pentasymmetron to the internal leaflet of the lipid membrane, may provide additional stabilization for the capsid. These results are consistent with the observations for other large, icosahedral dsDNA viruses that also utilize minor capsid proteins for stabilization and for determining their assembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1580.map.gz emd_1580.map.gz | 104.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1580-v30.xml emd-1580-v30.xml emd-1580.xml emd-1580.xml | 8.6 KB 8.6 KB | Display Display |  EMDB header EMDB header |
| Images |  1580.gif 1580.gif | 120 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1580 http://ftp.pdbj.org/pub/emdb/structures/EMD-1580 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1580 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1580 | HTTPS FTP |
-Validation report
| Summary document |  emd_1580_validation.pdf.gz emd_1580_validation.pdf.gz | 289.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1580_full_validation.pdf.gz emd_1580_full_validation.pdf.gz | 288.9 KB | Display | |
| Data in XML |  emd_1580_validation.xml.gz emd_1580_validation.xml.gz | 8.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1580 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1580 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1580 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1580 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1580.map.gz / Format: CCP4 / Size: 389.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1580.map.gz / Format: CCP4 / Size: 389.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Chilo Iridescent virus at 13 angstrom resolution | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.24 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
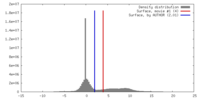
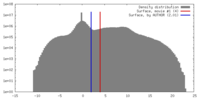
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : CIV
| Entire | Name: CIV |
|---|---|
| Components |
|
-Supramolecule #1000: CIV
| Supramolecule | Name: CIV / type: sample / ID: 1000 / Details: CIV virions / Number unique components: 1 |
|---|
-Supramolecule #1: Invertebrate iridescent virus 6
| Supramolecule | Name: Invertebrate iridescent virus 6 / type: virus / ID: 1 / Name.synonym: chilo iridescent virus / NCBI-ID: 176652 / Sci species name: Invertebrate iridescent virus 6 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: Yes / Virus empty: No / Syn species name: chilo iridescent virus |
|---|---|
| Host (natural) | Organism:  Lepidoptera (butterflies and moths) / synonym: INVERTEBRATES Lepidoptera (butterflies and moths) / synonym: INVERTEBRATES |
| Virus shell | Shell ID: 1 / Diameter: 1850 Å |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Grid | Details: holey carbon film |
|---|---|
| Vitrification | Cryogen name: NITROGEN / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: manual plunger |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM300FEG/T |
|---|---|
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7.0 µm / Number real images: 210 / Average electron dose: 22 e/Å2 / Bits/pixel: 12 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 33000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 33000 |
| Sample stage | Specimen holder: gatan 626 cryo holder / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: each micrograph |
|---|---|
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 13.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: AUTO3DEM / Number images used: 1800 |
| Final two d classification | Number classes: 1 |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)