[English] 日本語
 Yorodumi
Yorodumi- EMDB-15045: Localized reconstruction of bacteriophage phiCjT23 major capsid p... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Localized reconstruction of bacteriophage phiCjT23 major capsid protein trimer type 2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacteriophage / phiCjT23 / VIRUS | |||||||||
| Biological species |  Flavobacterium phage (virus) / unidentified (others) Flavobacterium phage (virus) / unidentified (others) | |||||||||
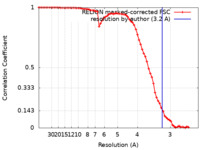
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Kejzar N / Abrishami V / Selvaraj M / Huiskonen JT | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Cryo-EM structure of ssDNA bacteriophage ΦCjT23 provides insight into early virus evolution. Authors: Nejc Kejzar / Elina Laanto / Ilona Rissanen / Vahid Abrishami / Muniyandi Selvaraj / Sylvain Moineau / Janne Ravantti / Lotta-Riina Sundberg / Juha T Huiskonen /    Abstract: The origin of viruses remains an open question. While lack of detectable sequence similarity hampers the analysis of distantly related viruses, structural biology investigations of conserved capsid ...The origin of viruses remains an open question. While lack of detectable sequence similarity hampers the analysis of distantly related viruses, structural biology investigations of conserved capsid protein structures facilitate the study of distant evolutionary relationships. Here we characterize the lipid-containing ssDNA temperate bacteriophage ΦCjT23, which infects Flavobacterium sp. (Bacteroidetes). We report ΦCjT23-like sequences in the genome of strains belonging to several Flavobacterium species. The virion structure determined by cryogenic electron microscopy reveals similarities to members of the viral kingdom Bamfordvirae that currently consists solely of dsDNA viruses with a major capsid protein composed of two upright β-sandwiches. The minimalistic structure of ΦCjT23 suggests that this phage serves as a model for the last common ancestor between ssDNA and dsDNA viruses in the Bamfordvirae. Both ΦCjT23 and the related phage FLiP infect Flavobacterium species found in several environments, suggesting that these types of viruses have a global distribution and a shared evolutionary origin. Detailed comparisons to related, more complex viruses not only expand our knowledge about this group of viruses but also provide a rare glimpse into early virus evolution. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15045.map.gz emd_15045.map.gz | 28.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15045-v30.xml emd-15045-v30.xml emd-15045.xml emd-15045.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15045_fsc.xml emd_15045_fsc.xml | 7.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_15045.png emd_15045.png | 105.8 KB | ||
| Masks |  emd_15045_msk_1.map emd_15045_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15045.cif.gz emd-15045.cif.gz | 5.1 KB | ||
| Others |  emd_15045_half_map_1.map.gz emd_15045_half_map_1.map.gz emd_15045_half_map_2.map.gz emd_15045_half_map_2.map.gz | 23.1 MB 23.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15045 http://ftp.pdbj.org/pub/emdb/structures/EMD-15045 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15045 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15045 | HTTPS FTP |
-Related structure data
| Related structure data |  8a02MC  7zzzC  8a01C  8a03C  8a04C  8a05C  8a06C M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15045.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15045.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.24 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15045_msk_1.map emd_15045_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_15045_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_15045_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : unidentified
| Entire | Name: unidentified (others) |
|---|---|
| Components |
|
-Supramolecule #1: unidentified
| Supramolecule | Name: unidentified / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 32644 / Sci species name: unidentified / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Flavobacterium johnsoniae UW101 (bacteria) Flavobacterium johnsoniae UW101 (bacteria) |
| Virus shell | Shell ID: 1 / Name: Capsid / Diameter: 600.0 Å / T number (triangulation number): 21 |
-Macromolecule #1: Major capsid protein P5
| Macromolecule | Name: Major capsid protein P5 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Flavobacterium phage (virus) Flavobacterium phage (virus) |
| Molecular weight | Theoretical: 26.180783 KDa |
| Sequence | String: MKIATITGVT KSPELQVTKA IGALILSSDV ALSALTTEKI SIYIERGNGS NVILANKVLL KDFILASTYG TENTQSDADN AMIALCELA DEGSIYLADK ESIKITLEDL ISDKRYDLHG IEEPQQTNNL FFFEQKSVAS EEFNKKIDVQ GFDLAIMTVD D SVSDLSYQ ...String: MKIATITGVT KSPELQVTKA IGALILSSDV ALSALTTEKI SIYIERGNGS NVILANKVLL KDFILASTYG TENTQSDADN AMIALCELA DEGSIYLADK ESIKITLEDL ISDKRYDLHG IEEPQQTNNL FFFEQKSVAS EEFNKKIDVQ GFDLAIMTVD D SVSDLSYQ YSNGQVVKYL PFELQTLSRD IDPIQAVLSD GKVVQGLTDR LTLPLVAVVG IEINKSQGSI INFVVRCLKT V |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 15.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.3 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8a02: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)