+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | FAP-80S Complex - Non-rotated state with empty A site | ||||||||||||
 Map data Map data | FAP-80S - Non-rotated state - Empty A site | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  | ||||||||||||
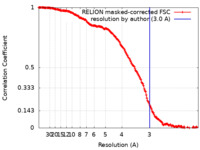
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | ||||||||||||
 Authors Authors | Ikeuchi K / Buschauer R / Berninghausen O / Becker T / Beckmann R | ||||||||||||
| Funding support |  Germany, European Union, 3 items Germany, European Union, 3 items
| ||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2022 Journal: Mol Cell / Year: 2022Title: Sensing of individual stalled 80S ribosomes by Fap1 for nonfunctional rRNA turnover. Authors: Sihan Li / Ken Ikeuchi / Misaki Kato / Robert Buschauer / Takato Sugiyama / Shungo Adachi / Hideo Kusano / Tohru Natsume / Otto Berninghausen / Yoshitaka Matsuo / Thomas Becker / Roland ...Authors: Sihan Li / Ken Ikeuchi / Misaki Kato / Robert Buschauer / Takato Sugiyama / Shungo Adachi / Hideo Kusano / Tohru Natsume / Otto Berninghausen / Yoshitaka Matsuo / Thomas Becker / Roland Beckmann / Toshifumi Inada /   Abstract: Cells can respond to stalled ribosomes by sensing ribosome collisions and employing quality control pathways. How ribosome stalling is resolved without collisions, however, has remained elusive. ...Cells can respond to stalled ribosomes by sensing ribosome collisions and employing quality control pathways. How ribosome stalling is resolved without collisions, however, has remained elusive. Here, focusing on noncolliding stalling exhibited by decoding-defective ribosomes, we identified Fap1 as a stalling sensor triggering 18S nonfunctional rRNA decay via polyubiquitination of uS3. Ribosome profiling revealed an enrichment of Fap1 at the translation initiation site but also an association with elongating individual ribosomes. Cryo-EM structures of Fap1-bound ribosomes elucidated Fap1 probing the mRNA simultaneously at both the entry and exit channels suggesting an mRNA stasis sensing activity, and Fap1 sterically hinders the formation of canonical collided di-ribosomes. Our findings indicate that individual stalled ribosomes are the potential signal for ribosome dysfunction, leading to accelerated turnover of the ribosome itself. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14992.map.gz emd_14992.map.gz | 241.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14992-v30.xml emd-14992-v30.xml emd-14992.xml emd-14992.xml | 24.4 KB 24.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14992_fsc.xml emd_14992_fsc.xml | 16.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_14992.png emd_14992.png | 165.1 KB | ||
| Others |  emd_14992_half_map_1.map.gz emd_14992_half_map_1.map.gz emd_14992_half_map_2.map.gz emd_14992_half_map_2.map.gz | 337.9 MB 338 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14992 http://ftp.pdbj.org/pub/emdb/structures/EMD-14992 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14992 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14992 | HTTPS FTP |
-Validation report
| Summary document |  emd_14992_validation.pdf.gz emd_14992_validation.pdf.gz | 802.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14992_full_validation.pdf.gz emd_14992_full_validation.pdf.gz | 801.9 KB | Display | |
| Data in XML |  emd_14992_validation.xml.gz emd_14992_validation.xml.gz | 23.7 KB | Display | |
| Data in CIF |  emd_14992_validation.cif.gz emd_14992_validation.cif.gz | 31.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14992 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14992 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14992 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14992 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14992.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14992.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | FAP-80S - Non-rotated state - Empty A site | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||
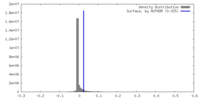
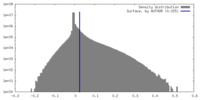
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map 1
| File | emd_14992_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
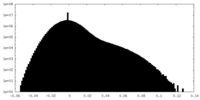
| Density Histograms |
-Half map: Half map 2
| File | emd_14992_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
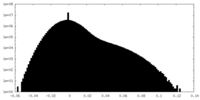
| Density Histograms |
- Sample components
Sample components
-Entire : Fap1-bound yeast 80S ribosome in non-rotated state with empty A site
| Entire | Name: Fap1-bound yeast 80S ribosome in non-rotated state with empty A site |
|---|---|
| Components |
|
-Supramolecule #1: Fap1-bound yeast 80S ribosome in non-rotated state with empty A site
| Supramolecule | Name: Fap1-bound yeast 80S ribosome in non-rotated state with empty A site type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#85 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 43.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)