[English] 日本語
 Yorodumi
Yorodumi- EMDB-14866: VelcroVax tandem HBcAg with SUMO-Affimer inserted at MIR (T=4 VLP) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | VelcroVax tandem HBcAg with SUMO-Affimer inserted at MIR (T=4 VLP) | |||||||||||||||
 Map data Map data | Density map filtered by local resolution. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | VelcroVax / Hepatitis B virus / Hepatitis B core antigen / Affimer / Vaccine / Recombinant / VLP / Antigen display / VIRUS LIKE PARTICLE | |||||||||||||||
| Biological species | synthetic construct (others) | |||||||||||||||
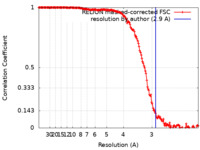
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||||||||
 Authors Authors | Kingston NJ / Grehan K / Snowden JS / Alzahrani J / Ranson NA / Rowlands DJ / Stonehouse NJ | |||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: mSphere / Year: 2023 Journal: mSphere / Year: 2023Title: VelcroVax: a "Bolt-On" Vaccine Platform for Glycoprotein Display. Authors: Natalie J Kingston / Keith Grehan / Joseph S Snowden / Mark Hassall / Jehad Alzahrani / Guido C Paesen / Lee Sherry / Connor Hayward / Amy Roe / Sam Stephen / Darren Tomlinson / Antra ...Authors: Natalie J Kingston / Keith Grehan / Joseph S Snowden / Mark Hassall / Jehad Alzahrani / Guido C Paesen / Lee Sherry / Connor Hayward / Amy Roe / Sam Stephen / Darren Tomlinson / Antra Zeltina / Katie J Doores / Neil A Ranson / Martin Stacey / Mark Page / Nicola J Rose / Thomas A Bowden / David J Rowlands / Nicola J Stonehouse /  Abstract: Having varied approaches to the design and manufacture of vaccines is critical in being able to respond to worldwide needs and newly emerging pathogens. Virus-like particles (VLPs) form the basis of ...Having varied approaches to the design and manufacture of vaccines is critical in being able to respond to worldwide needs and newly emerging pathogens. Virus-like particles (VLPs) form the basis of two of the most successful licensed vaccines (against hepatitis B virus [HBV] and human papillomavirus). They are produced by recombinant expression of viral structural proteins, which assemble into immunogenic nanoparticles. VLPs can be modified to present unrelated antigens, and here we describe a universal "bolt-on" platform (termed VelcroVax) where the capturing VLP and the target antigen are produced separately. We utilize a modified HBV core (HBcAg) VLP with surface expression of a high-affinity binding sequence (Affimer) directed against a SUMO tag and use this to capture SUMO-tagged gp1 glycoprotein from the arenavirus Junín virus (JUNV). Using this model system, we have solved the first high-resolution structures of VelcroVax VLPs and shown that the VelcroVax-JUNV gp1 complex induces superior humoral immune responses compared to the noncomplexed viral protein. We propose that this system could be modified to present a range of antigens and therefore form the foundation of future rapid-response vaccination strategies. The hepatitis B core protein (HBc) forms noninfectious virus-like particles, which can be modified to present a capturing molecule, allowing suitably tagged antigens to be bound on their surface. This system can be adapted and provides the foundation for a universal "bolt-on" vaccine platform (termed VelcroVax) that can be easily and rapidly modified to generate nanoparticle vaccine candidates. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14866.map.gz emd_14866.map.gz | 209.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14866-v30.xml emd-14866-v30.xml emd-14866.xml emd-14866.xml | 22.1 KB 22.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14866_fsc.xml emd_14866_fsc.xml | 16.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_14866.png emd_14866.png | 186.8 KB | ||
| Masks |  emd_14866_msk_1.map emd_14866_msk_1.map | 371.3 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14866.cif.gz emd-14866.cif.gz | 5.9 KB | ||
| Others |  emd_14866_additional_1.map.gz emd_14866_additional_1.map.gz emd_14866_additional_2.map.gz emd_14866_additional_2.map.gz emd_14866_additional_3.map.gz emd_14866_additional_3.map.gz emd_14866_half_map_1.map.gz emd_14866_half_map_1.map.gz emd_14866_half_map_2.map.gz emd_14866_half_map_2.map.gz | 291.4 MB 340.1 MB 67.1 MB 292.9 MB 292.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14866 http://ftp.pdbj.org/pub/emdb/structures/EMD-14866 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14866 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14866 | HTTPS FTP |
-Validation report
| Summary document |  emd_14866_validation.pdf.gz emd_14866_validation.pdf.gz | 937.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14866_full_validation.pdf.gz emd_14866_full_validation.pdf.gz | 937.3 KB | Display | |
| Data in XML |  emd_14866_validation.xml.gz emd_14866_validation.xml.gz | 23.7 KB | Display | |
| Data in CIF |  emd_14866_validation.cif.gz emd_14866_validation.cif.gz | 31.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14866 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14866 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14866 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14866 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14866.map.gz / Format: CCP4 / Size: 371.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14866.map.gz / Format: CCP4 / Size: 371.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density map filtered by local resolution. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.065 Å | ||||||||||||||||||||||||||||||||||||
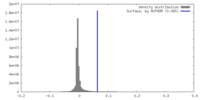
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
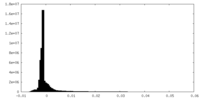
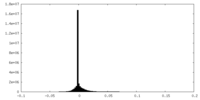
-Mask #1
| File |  emd_14866_msk_1.map emd_14866_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
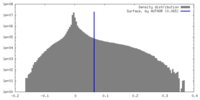
| Density Histograms |
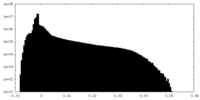
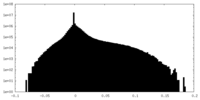
-Additional map: Unsharpened density map derived from 3D refinement.
| File | emd_14866_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened density map derived from 3D refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened density map.
| File | emd_14866_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened density map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened density map with mask applied to remove solvent.
| File | emd_14866_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened density map with mask applied to remove solvent. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
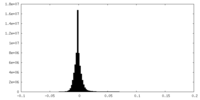
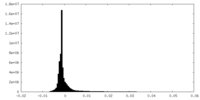
-Half map: Half map 1.
| File | emd_14866_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
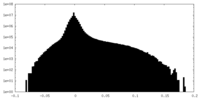
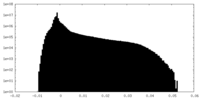
-Half map: Half map 2.
| File | emd_14866_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : T=4 virus-like particle for VelcroVax tandem HBcAg with SUMO-Affi...
| Entire | Name: T=4 virus-like particle for VelcroVax tandem HBcAg with SUMO-Affimer inserted at MIR |
|---|---|
| Components |
|
-Supramolecule #1: T=4 virus-like particle for VelcroVax tandem HBcAg with SUMO-Affi...
| Supramolecule | Name: T=4 virus-like particle for VelcroVax tandem HBcAg with SUMO-Affimer inserted at MIR type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: VelcroVax tandem HBcAg with SUMO-Affimer inserted at MIR
| Macromolecule | Name: VelcroVax tandem HBcAg with SUMO-Affimer inserted at MIR type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 52.79277 KDa |
| Recombinant expression | Organism:  Komagataella phaffii (fungus) Komagataella phaffii (fungus) |
| Sequence | String: MDIDPYKEFG ATVELLSFLP SDFFPSVRDL LDTASALYRE ALESPEHCSP HHTALRQAIL CWGELMTLAT WVGNNLEGSM ASAATGVRA VPGNENSLEI EELARFAVDE HNKKENALLE FVRVVKAKEQ IIIHENDADT MYYLTLEAKD GGKKKLYEAK V WVKGIMDG ...String: MDIDPYKEFG ATVELLSFLP SDFFPSVRDL LDTASALYRE ALESPEHCSP HHTALRQAIL CWGELMTLAT WVGNNLEGSM ASAATGVRA VPGNENSLEI EELARFAVDE HNKKENALLE FVRVVKAKEQ IIIHENDADT MYYLTLEAKD GGKKKLYEAK V WVKGIMDG LNKYNFKELQ EFKPVGDAGG RDPASRDLVV NYVNTNMGLK IRQLLWFHIS CLTFGRETVL EYLVSFGVWI RT PPAYRPP NAPILSTLPE TTVVGGSSGG SGGSGGSGGS GGSGGSTMDI DPYKEFGATV ELLSFLPSDF FPSVRDLLDT ASA LYREAL ESPEHCSPHH TALRQAILCW GELMTLATWV GNNLEFAGAS DPASRDLVVN YVNTNMGLKI RQLLWFHISC LTFG RETVL EYLVSFGVWI RTPPAYRPPN APILSTLPET TVVRRRDRGR SPRRRTPSPR RRRSQSPRRR RSQSRESQC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 60.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)