+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | AMC009 SOSIPv5.2 in complex with Fabs ACS114 and ACS122 | ||||||||||||
 Map data Map data | AMC009 SOSIPv5.2 ACS114 Fab ACS122 Fab | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | HIV-1 / antibodies / silent face / gp120-gp41 interface / VIRAL PROTEIN | ||||||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
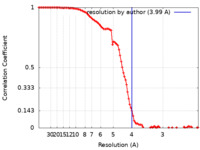
| Method | single particle reconstruction / cryo EM / Resolution: 3.99 Å | ||||||||||||
 Authors Authors | van Schooten J / Ozorowski G / Ward A | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2022 Journal: PLoS Pathog / Year: 2022Title: Complementary antibody lineages achieve neutralization breadth in an HIV-1 infected elite neutralizer. Authors: Jelle van Schooten / Anna Schorcht / Elinaz Farokhi / Jeffrey C Umotoy / Hongmei Gao / Tom L G M van den Kerkhof / Jessica Dorning / Tim G Rijkhold Meesters / Patricia van der Woude / Judith ...Authors: Jelle van Schooten / Anna Schorcht / Elinaz Farokhi / Jeffrey C Umotoy / Hongmei Gao / Tom L G M van den Kerkhof / Jessica Dorning / Tim G Rijkhold Meesters / Patricia van der Woude / Judith A Burger / Tom Bijl / Riham Ghalaiyini / Alba Torrents de la Peña / Hannah L Turner / Celia C Labranche / Robyn L Stanfield / Devin Sok / Hanneke Schuitemaker / David C Montefiori / Dennis R Burton / Gabriel Ozorowski / Michael S Seaman / Ian A Wilson / Rogier W Sanders / Andrew B Ward / Marit J van Gils /   Abstract: Broadly neutralizing antibodies (bNAbs) have remarkable breadth and potency against most HIV-1 subtypes and are able to prevent HIV-1 infection in animal models. However, bNAbs are extremely ...Broadly neutralizing antibodies (bNAbs) have remarkable breadth and potency against most HIV-1 subtypes and are able to prevent HIV-1 infection in animal models. However, bNAbs are extremely difficult to induce by vaccination. Defining the developmental pathways towards neutralization breadth can assist in the design of strategies to elicit protective bNAb responses by vaccination. Here, HIV-1 envelope glycoproteins (Env)-specific IgG+ B cells were isolated at various time points post infection from an HIV-1 infected elite neutralizer to obtain monoclonal antibodies (mAbs). Multiple antibody lineages were isolated targeting distinct epitopes on Env, including the gp120-gp41 interface, CD4-binding site, silent face and V3 region. The mAbs each neutralized a diverse set of HIV-1 strains from different clades indicating that the patient's remarkable serum breadth and potency might have been the result of a polyclonal mixture rather than a single bNAb lineage. High-resolution cryo-electron microscopy structures of the neutralizing mAbs (NAbs) in complex with an Env trimer generated from the same individual revealed that the NAbs used multiple strategies to neutralize the virus; blocking the receptor binding site, binding to HIV-1 Env N-linked glycans, and disassembly of the trimer. These results show that diverse NAbs can complement each other to achieve a broad and potent neutralizing serum response in HIV-1 infected individuals. Hence, the induction of combinations of moderately broad NAbs might be a viable vaccine strategy to protect against a wide range of circulating HIV-1 viruses. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14783.map.gz emd_14783.map.gz | 97.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14783-v30.xml emd-14783-v30.xml emd-14783.xml emd-14783.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14783_fsc.xml emd_14783_fsc.xml | 10.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_14783.png emd_14783.png | 84.6 KB | ||
| Masks |  emd_14783_msk_1.map emd_14783_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14783.cif.gz emd-14783.cif.gz | 6.9 KB | ||
| Others |  emd_14783_half_map_1.map.gz emd_14783_half_map_1.map.gz emd_14783_half_map_2.map.gz emd_14783_half_map_2.map.gz | 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14783 http://ftp.pdbj.org/pub/emdb/structures/EMD-14783 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14783 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14783 | HTTPS FTP |
-Validation report
| Summary document |  emd_14783_validation.pdf.gz emd_14783_validation.pdf.gz | 963.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14783_full_validation.pdf.gz emd_14783_full_validation.pdf.gz | 963.2 KB | Display | |
| Data in XML |  emd_14783_validation.xml.gz emd_14783_validation.xml.gz | 17.9 KB | Display | |
| Data in CIF |  emd_14783_validation.cif.gz emd_14783_validation.cif.gz | 23 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14783 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14783 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14783 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14783 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14783.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14783.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | AMC009 SOSIPv5.2 ACS114 Fab ACS122 Fab | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||
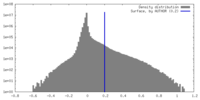
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14783_msk_1.map emd_14783_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: AMC009 SOSIPv5.2 ACS114 Fab ACS122 Fab half map A
| File | emd_14783_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | AMC009 SOSIPv5.2 ACS114 Fab ACS122 Fab half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: AMC009 SOSIPv5.2 ACS114 Fab ACS122 Fab half map B
| File | emd_14783_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | AMC009 SOSIPv5.2 ACS114 Fab ACS122 Fab half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : AMC009 SOSIPv5.2 in complex with Fabs ACS114 and ACS122
| Entire | Name: AMC009 SOSIPv5.2 in complex with Fabs ACS114 and ACS122 |
|---|---|
| Components |
|
-Supramolecule #1: AMC009 SOSIPv5.2 in complex with Fabs ACS114 and ACS122
| Supramolecule | Name: AMC009 SOSIPv5.2 in complex with Fabs ACS114 and ACS122 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17 kDa/nm |
-Macromolecule #1: AMC009 SOSIPv5.2 envelope glycoprotein gp120
| Macromolecule | Name: AMC009 SOSIPv5.2 envelope glycoprotein gp120 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 54.145488 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ARADKLWVTV YYGVPVWKEA TTTLFCASDA KAYDTEVRNV WATHCCVPTD PNPQEVVLEN VTENFNMWKN DMVEQMHEDI ISLWDQSLK PCVKLTPLCV TLNCTDYVGN ATNASTTNAT GGIGGTVERG EIKNCSFNIT TSIRDKVQKE YALFYKLDIV P IDNDNTNN ...String: ARADKLWVTV YYGVPVWKEA TTTLFCASDA KAYDTEVRNV WATHCCVPTD PNPQEVVLEN VTENFNMWKN DMVEQMHEDI ISLWDQSLK PCVKLTPLCV TLNCTDYVGN ATNASTTNAT GGIGGTVERG EIKNCSFNIT TSIRDKVQKE YALFYKLDIV P IDNDNTNN SYRLINCNTS VIKQACPKVS FEPIPIHYCA PAGFAILKCN DKKFNGTGPC TNVSTVQCTH GIRPVVSTQL LL NGSLAEK EVVIRSQNFT NNAKVIIVQL NESVVINCTR PNNNTRKSIH IAPGRWFYTT GAIIGDIRQA HCNISRVKWN NTL KQIATK LREQFKNKTI AFNQSSGGDP EIVMHSFNCG GEFFYCNTTQ LFNSTWNDTE VSNYNDITHI TLPCRIKQII NMWQ KVGKA MYAPPIRGQI RCSSNITGLL LTRDGGSNEN KTSETETFRP AGGDMRDNWR SELYKYKVVK IEPLGVAPTK CKRRV VQ |
-Macromolecule #2: AMC009 SOSIPv5.2 envelope glycoprotein gp41
| Macromolecule | Name: AMC009 SOSIPv5.2 envelope glycoprotein gp41 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.423865 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGAIGAVFL GFLGAAGSTM GAASMTLTVQ ARQLLSGIVQ QQNNLLRAPE CQQHMLKLTV WGIKQLQARV LAVERYLRDQ QLLGIWGCS GKLICCTAVP WNNSWSNRSL DMIWNNMTWI EWEREIDNYT GLIYNLLEES QNQQEKNEQE LLELD |
-Macromolecule #3: ACS114 heavy chain
| Macromolecule | Name: ACS114 heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.280985 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLQQWGGG LLKPSQTLSL TCAVYRWTFN DHYWSWVRQS PGKGLEWIGE ISWGGATNYN PSLKSRVTMS VDTSMSHVSL KMTSVTAAD TGVYYCVRVG PGPHMAALDY WGHGSRVLVS S |
-Macromolecule #4: ACS114 light chain
| Macromolecule | Name: ACS114 light chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.186664 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVMTQTPLS SPVTLGQPAS ISCGSSQSLV HSDGNTYLSW LQQRPGQPPR LLIYKISNRI SGLPDRFSGS GAETNFTLKI SRVEAEDVG LYYCVQGTQF PWTSGQGTKV EIK |
-Macromolecule #5: ACS122 heavy chain
| Macromolecule | Name: ACS122 heavy chain / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.774439 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLQESGPG LVKPSEALSL TCSVSGVAIS RHYWNWIRQP PGKGLEWIGY IFFNGNTNYS PSLKSRVTIS VDTSKNEFSL TLRSVTAAD TAVYYCAREK SVVEPDNMVR WFDPWGQGTL VTVSS |
-Macromolecule #6: ACS122 light chain
| Macromolecule | Name: ACS122 light chain / type: protein_or_peptide / ID: 6 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.668917 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SYELTQPPSV SVAPGKTARI TCGGNNIGSK SVHWYQQKPG QAPVLVIYYD SDRPSGIPER FSGSKSGNTA TLTISRVEAG DEADYYCQV WDSSRDHCVF GIGTKVTVL |
-Macromolecule #13: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 13 / Number of copies: 21 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 49.3 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.7 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)