+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of the vacant Candida albicans 80S ribosome | ||||||||||||||||||
 Map data Map data | Cryo-EM map of the vacant Candida albicans 80S ribosome | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationfilamentous growth of a population of unicellular organisms in response to neutral pH / cellular response to neutral pH / negative regulation of cell integrity MAPK cascade / positive regulation of conjugation with cellular fusion / filamentous growth of a population of unicellular organisms in response to biotic stimulus / filamentous growth of a population of unicellular organisms in response to starvation / yeast-form cell wall / GCN2-mediated signaling / regulation of cytoplasmic translation / fungal biofilm matrix ...filamentous growth of a population of unicellular organisms in response to neutral pH / cellular response to neutral pH / negative regulation of cell integrity MAPK cascade / positive regulation of conjugation with cellular fusion / filamentous growth of a population of unicellular organisms in response to biotic stimulus / filamentous growth of a population of unicellular organisms in response to starvation / yeast-form cell wall / GCN2-mediated signaling / regulation of cytoplasmic translation / fungal biofilm matrix / hyphal cell wall / invasive growth in response to glucose limitation / triplex DNA binding / negative regulation of p38MAPK cascade / ribosome hibernation / translation elongation factor binding / regulation of translational initiation in response to stress / filamentous growth / negative regulation of glucose mediated signaling pathway / negative regulation of translational frameshifting / positive regulation of translational fidelity / ribosome-associated ubiquitin-dependent protein catabolic process / GDP-dissociation inhibitor activity / pre-mRNA 5'-splice site binding / preribosome, small subunit precursor / telomeric DNA binding / mRNA destabilization / negative regulation of mRNA splicing, via spliceosome / translational elongation / G-protein alpha-subunit binding / TOR signaling / endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / protein-RNA complex assembly / ribosomal subunit export from nucleus / ribosomal small subunit export from nucleus / positive regulation of autophagy / translation regulator activity / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / translation repressor activity / DNA-(apurinic or apyrimidinic site) endonuclease activity / maturation of LSU-rRNA / telomere maintenance / cellular response to starvation / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / rescue of stalled ribosome / ribosomal large subunit biogenesis / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA / small-subunit processome / protein kinase C binding / maintenance of translational fidelity / modification-dependent protein catabolic process / protein tag activity / rRNA processing / ribosomal small subunit biogenesis / small ribosomal subunit rRNA binding / extracellular vesicle / ribosome biogenesis / ribosome binding / ribosomal small subunit assembly / small ribosomal subunit / 5S rRNA binding / large ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / ribosomal large subunit assembly / cytoplasmic translation / cytosolic large ribosomal subunit / negative regulation of translation / cell adhesion / rRNA binding / ribosome / protein ubiquitination / structural constituent of ribosome / ribonucleoprotein complex / positive regulation of protein phosphorylation / G protein-coupled receptor signaling pathway / translation / mRNA binding / ubiquitin protein ligase binding / negative regulation of apoptotic process / nucleolus / cell surface / RNA binding / zinc ion binding / extracellular region / membrane / nucleus / metal ion binding / cytosol / cytoplasm Similarity search - Function | ||||||||||||||||||
| Biological species |  Candida albicans SC5314 (yeast) Candida albicans SC5314 (yeast) | ||||||||||||||||||
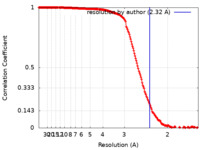
| Method | single particle reconstruction / cryo EM / Resolution: 2.32 Å | ||||||||||||||||||
 Authors Authors | Zgadzay Y / Kolosova O / Stetsenko A / Jenner L / Guskov A / Yusupova G / Yusupov M | ||||||||||||||||||
| Funding support |  France, 5 items France, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: E-site drug specificity of the human pathogen ribosome. Authors: Yury Zgadzay / Olga Kolosova / Artem Stetsenko / Cheng Wu / David Bruchlen / Konstantin Usachev / Shamil Validov / Lasse Jenner / Andrey Rogachev / Gulnara Yusupova / Matthew S Sachs / ...Authors: Yury Zgadzay / Olga Kolosova / Artem Stetsenko / Cheng Wu / David Bruchlen / Konstantin Usachev / Shamil Validov / Lasse Jenner / Andrey Rogachev / Gulnara Yusupova / Matthew S Sachs / Albert Guskov / Marat Yusupov /     Abstract: is a widespread commensal fungus with substantial pathogenic potential and steadily increasing resistance to current antifungal drugs. It is known to be resistant to cycloheximide (CHX) that binds ... is a widespread commensal fungus with substantial pathogenic potential and steadily increasing resistance to current antifungal drugs. It is known to be resistant to cycloheximide (CHX) that binds to the E-transfer RNA binding site of the ribosome. Because of lack of structural information, it is neither possible to understand the nature of the resistance nor to develop novel inhibitors. To overcome this issue, we determined the structure of the vacant 80 ribosome at 2.3 angstroms and its complexes with bound inhibitors at resolutions better than 2.9 angstroms using cryo-electron microscopy. Our structures reveal how a change in a conserved amino acid in ribosomal protein eL42 explains CHX resistance in and forms a basis for further antifungal drug development. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13737.map.gz emd_13737.map.gz | 478.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13737-v30.xml emd-13737-v30.xml emd-13737.xml emd-13737.xml | 99.3 KB 99.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13737_fsc.xml emd_13737_fsc.xml | 17.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_13737.png emd_13737.png | 238.4 KB | ||
| Others |  emd_13737_additional_1.map.gz emd_13737_additional_1.map.gz emd_13737_additional_2.map.gz emd_13737_additional_2.map.gz | 245.5 MB 243.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13737 http://ftp.pdbj.org/pub/emdb/structures/EMD-13737 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13737 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13737 | HTTPS FTP |
-Validation report
| Summary document |  emd_13737_validation.pdf.gz emd_13737_validation.pdf.gz | 533.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13737_full_validation.pdf.gz emd_13737_full_validation.pdf.gz | 533.1 KB | Display | |
| Data in XML |  emd_13737_validation.xml.gz emd_13737_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  emd_13737_validation.cif.gz emd_13737_validation.cif.gz | 22.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13737 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13737 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13737 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13737 | HTTPS FTP |
-Related structure data
| Related structure data |  7pzyMC  7q08C  7q0fC  7q0pC  7q0rC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13737.map.gz / Format: CCP4 / Size: 506 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13737.map.gz / Format: CCP4 / Size: 506 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of the vacant Candida albicans 80S ribosome | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.836 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Focused cryo-EM map of the vacant Candida albicans...
| File | emd_13737_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused cryo-EM map of the vacant Candida albicans 80S ribosome (on the head of 40S subunit) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Focused cryo-EM map of the vacant Candida albicans...
| File | emd_13737_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused cryo-EM map of the vacant Candida albicans 80S ribosome (on the 40S subunit) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Candida albicans 80S ribosome
+Supramolecule #1: Candida albicans 80S ribosome
+Macromolecule #1: 25S ribosomal RNA
+Macromolecule #2: 5S ribosomal RNA
+Macromolecule #3: 5.8S ribosomal RNA
+Macromolecule #4: Mixture of endogenous E-tRNAs
+Macromolecule #47: 18S ribosomal RNA
+Macromolecule #5: Ribosomal 60S subunit protein L2A
+Macromolecule #6: 60S ribosomal protein L3
+Macromolecule #7: Ribosomal 60S subunit protein L4B
+Macromolecule #8: Ribosomal 60S subunit protein L5
+Macromolecule #9: 60S ribosomal protein L6
+Macromolecule #10: Ribosomal 60S subunit protein L7A
+Macromolecule #11: 60S ribosomal protein L8
+Macromolecule #12: Ribosomal 60S subunit protein L9B
+Macromolecule #13: Ribosomal 60S subunit protein L10
+Macromolecule #14: Ribosomal 60S subunit protein L11B
+Macromolecule #15: 60S ribosomal protein L13
+Macromolecule #16: Ribosomal 60S subunit protein L14B
+Macromolecule #17: Ribosomal protein L15
+Macromolecule #18: Ribosomal 60S subunit protein L16A
+Macromolecule #19: Ribosomal 60S subunit protein L17B
+Macromolecule #20: Ribosomal 60S subunit protein L18A
+Macromolecule #21: Ribosomal protein L19
+Macromolecule #22: 60S ribosomal protein L20
+Macromolecule #23: Ribosomal 60S subunit protein L21A
+Macromolecule #24: Ribosomal 60S subunit protein L22B
+Macromolecule #25: Ribosomal 60S subunit protein L23B
+Macromolecule #26: Ribosomal 60S subunit protein L24A
+Macromolecule #27: Ribosomal 60S subunit protein L25
+Macromolecule #28: Ribosomal 60S subunit protein L26B
+Macromolecule #29: 60S ribosomal protein L27
+Macromolecule #30: Ribosomal 60S subunit protein L28
+Macromolecule #31: 60S ribosomal protein L29
+Macromolecule #32: Ribosomal 60S subunit protein L30
+Macromolecule #33: Ribosomal 60S subunit protein L31B
+Macromolecule #34: Ribosomal 60S subunit protein L32
+Macromolecule #35: Ribosomal 60S subunit protein L33A
+Macromolecule #36: Ribosomal 60S subunit protein L34B
+Macromolecule #37: Ribosomal 60S subunit protein L35A
+Macromolecule #38: 60S ribosomal protein L36
+Macromolecule #39: Ribosomal protein L37
+Macromolecule #40: Ribosomal 60S subunit protein L38
+Macromolecule #41: 60S ribosomal protein L39
+Macromolecule #42: Rpl40bp
+Macromolecule #43: 60S ribosomal protein L41
+Macromolecule #44: Ribosomal 60S subunit protein L42A
+Macromolecule #45: Ribosomal 60S subunit protein L43A
+Macromolecule #46: HABP4_PAI-RBP1 domain-containing protein
+Macromolecule #48: 40S ribosomal protein S0
+Macromolecule #49: 40S ribosomal protein S1
+Macromolecule #50: Ribosomal 40S subunit protein S2
+Macromolecule #51: Ribosomal 40S subunit protein S3
+Macromolecule #52: 40S ribosomal protein S4
+Macromolecule #53: Ribosomal 40S subunit protein S5
+Macromolecule #54: 40S ribosomal protein S6
+Macromolecule #55: 40S ribosomal protein S7
+Macromolecule #56: 40S ribosomal protein S8
+Macromolecule #57: Ribosomal 40S subunit protein S9B
+Macromolecule #58: Ribosomal 40S subunit protein S10A
+Macromolecule #59: Ribosomal 40S subunit protein S11A
+Macromolecule #60: 40S ribosomal protein S12
+Macromolecule #61: Ribosomal 40S subunit protein S13
+Macromolecule #62: Ribosomal 40S subunit protein S14B
+Macromolecule #63: Ribosomal 40S subunit protein S15
+Macromolecule #64: Ribosomal 40S subunit protein S16A
+Macromolecule #65: Ribosomal 40S subunit protein S17B
+Macromolecule #66: Ribosomal 40S subunit protein S18B
+Macromolecule #67: Ribosomal 40S subunit protein S19A
+Macromolecule #68: Ribosomal 40S subunit protein S20
+Macromolecule #69: 40S ribosomal protein S21
+Macromolecule #70: 40S ribosomal protein S22-A
+Macromolecule #71: Ribosomal 40S subunit protein S23B
+Macromolecule #72: 40S ribosomal protein S24
+Macromolecule #73: 40S ribosomal protein S25
+Macromolecule #74: 40S ribosomal protein S26
+Macromolecule #75: 40S ribosomal protein S27
+Macromolecule #76: Ribosomal 40S subunit protein S28B
+Macromolecule #77: Ribosomal 40S subunit protein S29A
+Macromolecule #78: 40S ribosomal protein S30
+Macromolecule #79: Ubiquitin-ribosomal 40S subunit protein S31 fusion protein
+Macromolecule #80: Guanine nucleotide-binding protein subunit beta-like protein
+Macromolecule #81: SPERMIDINE
+Macromolecule #82: 1,4-DIAMINOBUTANE
+Macromolecule #83: MAGNESIUM ION
+Macromolecule #84: ZINC ION
+Macromolecule #85: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: OTHER / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.9 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)