+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13642 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Alpha-latrocrustotoxin monomer | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Pore-forming toxin / TOXIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationother organism cell membrane / host cell presynaptic membrane / exocytosis / toxin activity / extracellular region Similarity search - Function | ||||||||||||
| Biological species |  Latrodectus tredecimguttatus (black widow) Latrodectus tredecimguttatus (black widow) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.03 Å | ||||||||||||
 Authors Authors | Chen M / Gatsogiannis C | ||||||||||||
| Funding support |  Japan, 3 items Japan, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Molecular architecture of black widow spider neurotoxins. Authors: Minghao Chen / Daniel Blum / Lena Engelhard / Stefan Raunser / Richard Wagner / Christos Gatsogiannis /  Abstract: Latrotoxins (LaTXs) are presynaptic pore-forming neurotoxins found in the venom of Latrodectus spiders. The venom contains a toxic cocktail of seven LaTXs, with one of them targeting vertebrates (α- ...Latrotoxins (LaTXs) are presynaptic pore-forming neurotoxins found in the venom of Latrodectus spiders. The venom contains a toxic cocktail of seven LaTXs, with one of them targeting vertebrates (α-latrotoxin (α-LTX)), five specialized on insects (α, β, γ, δ, ε- latroinsectotoxins (LITs), and one on crustaceans (α-latrocrustatoxin (α-LCT)). LaTXs bind to specific receptors on the surface of neuronal cells, inducing the release of neurotransmitters either by directly stimulating exocytosis or by forming Ca-conductive tetrameric pores in the membrane. Despite extensive studies in the past decades, a high-resolution structure of a LaTX is not yet available and the precise mechanism of LaTX action remains unclear. Here, we report cryoEM structures of the α-LCT monomer and the δ-LIT dimer. The structures reveal that LaTXs are organized in four domains. A C-terminal domain of ankyrin-like repeats shields a central membrane insertion domain of six parallel α-helices. Both domains are flexibly linked via an N-terminal α-helical domain and a small β-sheet domain. A comparison between the structures suggests that oligomerization involves major conformational changes in LaTXs with longer C-terminal domains. Based on our data we propose a cyclic mechanism of oligomerization, taking place prior membrane insertion. Both recombinant α-LCT and δ-LIT form channels in artificial membrane bilayers, that are stabilized by Ca ions and allow calcium flux at negative membrane potentials. Our comparative analysis between α-LCT and δ-LIT provides first crucial insights towards understanding the molecular mechanism of the LaTX family. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13642.map.gz emd_13642.map.gz | 2.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13642-v30.xml emd-13642-v30.xml emd-13642.xml emd-13642.xml | 12.4 KB 12.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13642.png emd_13642.png | 69.7 KB | ||
| Filedesc metadata |  emd-13642.cif.gz emd-13642.cif.gz | 6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13642 http://ftp.pdbj.org/pub/emdb/structures/EMD-13642 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13642 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13642 | HTTPS FTP |
-Related structure data
| Related structure data |  7ptxMC  7ptyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10827 (Title: CryoEM single particle dataset of alpha-latrocrustotoxin monomer EMPIAR-10827 (Title: CryoEM single particle dataset of alpha-latrocrustotoxin monomerData size: 5.1 TB Data #1: Raw images of the alpha-latrocrustatoxin monomer part1 [micrographs - multiframe] Data #2: Raw images of the alpha-latrocrustatoxin monomer part2 [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13642.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13642.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.9 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
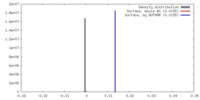
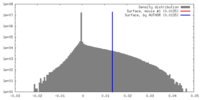
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Alpha-latrocrustotoxin-Lt1a in soluble monomeric state
| Entire | Name: Alpha-latrocrustotoxin-Lt1a in soluble monomeric state |
|---|---|
| Components |
|
-Supramolecule #1: Alpha-latrocrustotoxin-Lt1a in soluble monomeric state
| Supramolecule | Name: Alpha-latrocrustotoxin-Lt1a in soluble monomeric state type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Latrodectus tredecimguttatus (black widow) Latrodectus tredecimguttatus (black widow) |
| Molecular weight | Theoretical: 139.8 KDa |
-Macromolecule #1: Alpha-latrocrustotoxin-Lt1a
| Macromolecule | Name: Alpha-latrocrustotoxin-Lt1a / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Latrodectus tredecimguttatus (black widow) Latrodectus tredecimguttatus (black widow) |
| Molecular weight | Theoretical: 140.010531 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MWSHPQFEKG SAGSAAGSGA GWSHPQFEKG AGLEVLFQGP NSEMKGKRVI SKREMSKADQ CTFLSYQSVA YGTLGDVAGD VSSIEGADL VATPIAAGGH LAKGATDAAM IAMDCSSIPF DEIKQQLNQR FNEVDKKLQK GAEALENVTE LAEKTYSSVE K MRVEMREG ...String: MWSHPQFEKG SAGSAAGSGA GWSHPQFEKG AGLEVLFQGP NSEMKGKRVI SKREMSKADQ CTFLSYQSVA YGTLGDVAGD VSSIEGADL VATPIAAGGH LAKGATDAAM IAMDCSSIPF DEIKQQLNQR FNEVDKKLQK GAEALENVTE LAEKTYSSVE K MRVEMREG FNHVIATIEN ANTKQIITGI NQIIQYFNDE RENINNRQKE DYVAKLQEPA SGNFLLYLRK SRTSEDGSLH SL LFKIINQ ELAIPNNAAD NNAIRALFAL FYGTQTFISI MFYLVKQYSY LADYHYQNGN LAEFNSNFDH MKTVFQDFKF TLI GINTSN SKPLVNTVLS IIEDVKNKRF IRNLRSNLYQ KIIKSTKSLL DLREKITKMD LPIIEDTPKS SVLINFREKS SSVP RIETP ILKWTPGTVV KYAIQYEQDG KYSKISKWSN PITVQRLANP YITIDKDRRN RLVFRQFGNE KPELISILDS SQNEF RDIH RDLYNAAQMP YKETALGICR KLIDSGAQVG ASFEMGRKSI HASATAGNDD VARLLLAKNN GLLNVPDKNG YTPLHI ASE RKNNDFVKFL LEKGADVNVR TFANELTPLH LAARQDFTII VKTLMEKRGI DVNAKERAGF TPLHLSITSN SRAARTL IN ETPAGINIKS NSGLTPLHLA VLQNNLSAAK VLVKSNKKVK LNEMDNNGMT PLHYASMLGN LEFVKYFTSE QGIDVNAK T KVKNWTPLHL AILFKKFDVA QSLLQVRNID ISTRADQAIT PLHLAAATGN SQIVKTILNS GAVVDQETAN GFTALHLAI MNPNTETPQF LIAKGANINA KTNDGSTPLH FAAALGKTNI FQLLMDKGAN IKAENLINQM PIHEAVVNGH LAIVKMLIEQ DSSLMNAKN MRDEYPFYLA AEKRYKDVFN YLESKGADVN EKNNDGNTLL HLFSINGEVE VVQFLIQNGA DFRLRNKERK S FFDLAVEF GHAGIVGYAI EENKVDLQEP YRGKTILYHA ICDSVKYDRI EVVRYFVETL NEDQCSPLQE AAAYAHLDLV KY FVQERGI NPTAFNNDNQ VSPLCIAIVG APCGFVKSCD TPERLDVVEY LVDKTPDINK ECDTQQSTPV SSAVYGNKVS ILN YLIRNG ADPNKKVRGD PPLFIAAMIG QYDIVKSLVE QHKIDVNTRN KEQFTPLHAA ASNDHIDVVK YLIQKGADVN AKGD ENLKP IDLAGEKSKA YLRSLGRRFF RNESPSKSFE ISSGLVPRGS HHHHHHHH UniProtKB: Alpha-latrocrustotoxin-Lt1a |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 1 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 69.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7ptx: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)