[English] 日本語
 Yorodumi
Yorodumi- EMDB-13510: Cryo-EM structure of the actomyosin-V complex in the rigor state ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13510 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the actomyosin-V complex in the rigor state (central 1er, young JASP-stabilized F-actin, class 4) | ||||||||||||||||||
 Map data Map data | Sharpened map of the central actomyosin-V-LC molecule filtered to local resolution (class 4) | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationminus-end directed microfilament motor activity / unconventional myosin complex / insulin-responsive compartment / muscle myosin complex / muscle filament sliding / myosin complex / myosin II complex / structural constituent of muscle / cytoskeletal motor activator activity / myosin heavy chain binding ...minus-end directed microfilament motor activity / unconventional myosin complex / insulin-responsive compartment / muscle myosin complex / muscle filament sliding / myosin complex / myosin II complex / structural constituent of muscle / cytoskeletal motor activator activity / myosin heavy chain binding / microfilament motor activity / tropomyosin binding / actin filament bundle / troponin I binding / filamentous actin / mesenchyme migration / cytoskeletal motor activity / skeletal muscle myofibril / actin filament bundle assembly / striated muscle thin filament / skeletal muscle thin filament assembly / actin monomer binding / Smooth Muscle Contraction / skeletal muscle tissue development / skeletal muscle fiber development / stress fiber / vesicle-mediated transport / titin binding / actin filament polymerization / muscle contraction / actin filament organization / protein localization to plasma membrane / actin filament / filopodium / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / cellular response to insulin stimulus / calcium-dependent protein binding / actin filament binding / actin cytoskeleton / lamellipodium / cell body / calmodulin binding / Golgi membrane / protein domain specific binding / hydrolase activity / calcium ion binding / positive regulation of gene expression / magnesium ion binding / ATP hydrolysis activity / extracellular exosome / ATP binding / identical protein binding / membrane / cytoplasm / cytosol Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   | ||||||||||||||||||
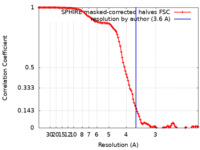
| Method | helical reconstruction / cryo EM / Resolution: 3.6 Å | ||||||||||||||||||
 Authors Authors | Pospich S / Sweeney HL / Houdusse A / Raunser S | ||||||||||||||||||
| Funding support |  Germany, European Union, Germany, European Union,  France, France,  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism. Authors: Sabrina Pospich / H Lee Sweeney / Anne Houdusse / Stefan Raunser /    Abstract: The molecular motor myosin undergoes a series of major structural transitions during its force-producing motor cycle. The underlying mechanism and its coupling to ATP hydrolysis and actin binding are ...The molecular motor myosin undergoes a series of major structural transitions during its force-producing motor cycle. The underlying mechanism and its coupling to ATP hydrolysis and actin binding are only partially understood, mostly due to sparse structural data on actin-bound states of myosin. Here, we report 26 high-resolution cryo-EM structures of the actomyosin-V complex in the strong-ADP, rigor, and a previously unseen post-rigor transition state that binds the ATP analog AppNHp. The structures reveal a high flexibility of myosin in each state and provide valuable insights into the structural transitions of myosin-V upon ADP release and binding of AppNHp, as well as the actomyosin interface. In addition, they show how myosin is able to specifically alter the structure of F-actin. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13510.map.gz emd_13510.map.gz | 1.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13510-v30.xml emd-13510-v30.xml emd-13510.xml emd-13510.xml | 26 KB 26 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13510_fsc.xml emd_13510_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_13510.png emd_13510.png | 94.4 KB | ||
| Masks |  emd_13510_msk_1.map emd_13510_msk_1.map | 125 MB |  Mask map Mask map | |
| Others |  emd_13510_additional_1.map.gz emd_13510_additional_1.map.gz emd_13510_additional_2.map.gz emd_13510_additional_2.map.gz emd_13510_half_map_1.map.gz emd_13510_half_map_1.map.gz emd_13510_half_map_2.map.gz emd_13510_half_map_2.map.gz | 1.3 MB 11.1 MB 59.9 MB 59.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13510 http://ftp.pdbj.org/pub/emdb/structures/EMD-13510 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13510 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13510 | HTTPS FTP |
-Related structure data
| Related structure data |  7pm2MC  7pltC  7pluC  7plvC  7plwC  7plxC  7plyC  7plzC  7pm0C  7pm1C  7pm3C  7pm5C  7pm6C  7pm7C  7pm8C  7pm9C  7pmaC  7pmbC  7pmcC  7pmdC  7pmeC  7pmfC  7pmgC  7pmhC  7pmiC  7pmjC  7pmlC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13510.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13510.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of the central actomyosin-V-LC molecule filtered to local resolution (class 4) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
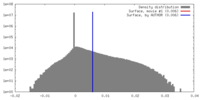
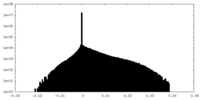
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13510_msk_1.map emd_13510_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened map of the central actomyosin-V-LC molecule filtered...
| File | emd_13510_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of the central actomyosin-V-LC molecule filtered to nominal resolution (class 4) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Denoised map of the central actomyosin-V-LC molecule (LAFTER,...
| File | emd_13510_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Denoised map of the central actomyosin-V-LC molecule (LAFTER, class 4) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map (signal subtracted particles, class 4)
| File | emd_13510_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map (signal subtracted particles, class 4) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map (signal subtracted particles, class 4)
| File | emd_13510_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map (signal subtracted particles, class 4) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Young actomyosin-V complex in the rigor state
| Entire | Name: Young actomyosin-V complex in the rigor state |
|---|---|
| Components |
|
-Supramolecule #1: Young actomyosin-V complex in the rigor state
| Supramolecule | Name: Young actomyosin-V complex in the rigor state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: Young ADP-Pi-bound F-actin stabilized with jasplakinolide and decorated with myosin-Va-LC in the rigor state (nucleotide-free) |
|---|
-Macromolecule #1: Myosin light chain 6B
| Macromolecule | Name: Myosin light chain 6B / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 17.090277 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIEFNKDQLE EFKEAFELFD RVGDGKILYS QCGDVMRALG QNPTNAEVLK VLGNPKSDEL KSRRVDFETF LPMLQAVAKN RGQGTYEDY LEGFRVFDKE GNGKVMGAEL RHVLTTLGEK MTEEEVETVL AGHEDSNGCI NYEAFLKHIL SV |
-Macromolecule #2: Unconventional myosin-Va
| Macromolecule | Name: Unconventional myosin-Va / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 91.363953 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAASELYTKY ARVWIPDPEE VWKSAELLKD YKPGDKVLQL RLEEGKDLEY CLDPKTKELP PLRNPDILVG ENDLTALSYL HEPAVLHNL KVRFIDSKLI YTYCGIVLVA INPYEQLPIY GEDIINAYSG QNMGDMDPHI FAVAEEAYKQ MARDERNQSI I VSGESGAG ...String: MAASELYTKY ARVWIPDPEE VWKSAELLKD YKPGDKVLQL RLEEGKDLEY CLDPKTKELP PLRNPDILVG ENDLTALSYL HEPAVLHNL KVRFIDSKLI YTYCGIVLVA INPYEQLPIY GEDIINAYSG QNMGDMDPHI FAVAEEAYKQ MARDERNQSI I VSGESGAG KTVSAKYAMR YFATVSGSAS EANVEEKVLA SNPIMESIGN AKTTRNDNSS RFGKYIEIGF DKRYRIIGAN MR TYLLEKS RVVFQAEEER NYHIFYQLCA SAALPEFKTL RLGNANYFHY TKQGGSPVID GIDDAKEMVN TRQACTLLGI SDS YQMGIF RILAGILHLG NVEFASRDSD SCAIPPKHDP LTIFCDLMGV DYEEMAHWLC HRKLATATET YIKPISKLHA INAR DALAK HIYANLFNWI VDHVNKALHS TVKQHSFIGV LDIYGFETFE INSFEQFCIN YANEKLQQQF NMHVFKLEQE EYMKE QIPW TLIDFYDNQP CINLIEAKMG VLDLLDEECK MPKGSDDTWA QKLYNTHLNK CALFEKPRLS NKAFIIKHFA DKVEYQ CEG FLEKNKDTVY EEQIKVLKSS KKFKLLPELF QDEEKAISPT SATPSGRVPL SRTPVKPAKA RPGQTSKEHK KTVGHQF RN SLHLLMETLN ATTPHYVRCI KPNDFKFPFT FDEKRAVQQL RACGVLETIR ISAAGFPSRW TYQEFFSRYR VLMKQKDV L SDRKQTCKNV LEKLILDKDK YQFGKTKIFF RAGQVAYLEK IRADKLRAAC IRIQKTIRGW LMRKKYMRMR R |
-Macromolecule #3: Actin, alpha skeletal muscle
| Macromolecule | Name: Actin, alpha skeletal muscle / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 42.109973 KDa |
| Sequence | String: MCDEDETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGVMVGMG QKDSYVGDEA QSKRGILTLK YPIE(HIC)G IIT NWDDMEKIWH HTFYNELRVA PEEHPTLLTE APLNPKANRE KMTQIMFETF NVPAMYVAIQ AVLSLYASGR TTGIVLD SG DGVTHNVPIY ...String: MCDEDETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGVMVGMG QKDSYVGDEA QSKRGILTLK YPIE(HIC)G IIT NWDDMEKIWH HTFYNELRVA PEEHPTLLTE APLNPKANRE KMTQIMFETF NVPAMYVAIQ AVLSLYASGR TTGIVLD SG DGVTHNVPIY EGYALPHAIM RLDLAGRDLT DYLMKILTER GYSFVTTAER EIVRDIKEKL CYVALDFENE MATAASSS S LEKSYELPDG QVITIGNERF RCPETLFQPS FIGMESAGIH ETTYNSIMKC DIDIRKDLYA NNVMSGGTTM YPGIADRMQ KEITALAPST MKIKIIAPPE RKYSVWIGGS ILASLSTFQQ MWITKQEYDE AGPSIVHRKC F |
-Macromolecule #4: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 1 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #5: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #7: Jasplakinolide
| Macromolecule | Name: Jasplakinolide / type: ligand / ID: 7 / Number of copies: 1 / Formula: 9UE |
|---|---|
| Molecular weight | Theoretical: 709.67 Da |
| Chemical component information |  ChemComp-9UE: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 286 K / Instrument: FEI VITROBOT MARK III / Details: On grid decoration. |
| Details | Signal subtracted |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Spherical aberration corrector: Cs-corrected microscope / Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Number real images: 3336 / Average exposure time: 15.0 sec. / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 0.0 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)