[English] 日本語
 Yorodumi
Yorodumi- EMDB-13034: Cryo-EM structure of the cellular negative regulator TFS4 bound t... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13034 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the cellular negative regulator TFS4 bound to the archaeal RNA polymerase | |||||||||
 Map data Map data | Sharpened map from Autosharpen, Phenix v1.15.2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | negative regulator / inhibitor / Archaea / Sso TFS4 / trancript cleavage factor / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology information3 iron, 4 sulfur cluster binding / transcription elongation by RNA polymerase I / tRNA transcription by RNA polymerase III / transcription by RNA polymerase I / DNA-directed RNA polymerase complex / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / defense response to virus ...3 iron, 4 sulfur cluster binding / transcription elongation by RNA polymerase I / tRNA transcription by RNA polymerase III / transcription by RNA polymerase I / DNA-directed RNA polymerase complex / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / defense response to virus / transcription by RNA polymerase II / protein dimerization activity / nucleotide binding / negative regulation of DNA-templated transcription / DNA-templated transcription / magnesium ion binding / DNA binding / zinc ion binding / metal ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Sulfolobus acidocaldarius DSM 639 (acidophilic) / Sulfolobus acidocaldarius DSM 639 (acidophilic) /   Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) (archaea) Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) (archaea) | |||||||||
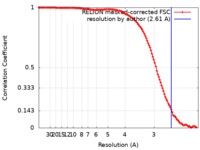
| Method | single particle reconstruction / cryo EM / Resolution: 2.61 Å | |||||||||
 Authors Authors | Pilotto S / Fouqueau T | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structural basis of RNA polymerase inhibition by viral and host factors. Authors: Simona Pilotto / Thomas Fouqueau / Natalya Lukoyanova / Carol Sheppard / Soizick Lucas-Staat / Luis Miguel Díaz-Santín / Dorota Matelska / David Prangishvili / Alan C M Cheung / Finn Werner /   Abstract: RNA polymerase inhibition plays an important role in the regulation of transcription in response to environmental changes and in the virus-host relationship. Here we present the high-resolution ...RNA polymerase inhibition plays an important role in the regulation of transcription in response to environmental changes and in the virus-host relationship. Here we present the high-resolution structures of two such RNAP-inhibitor complexes that provide the structural bases underlying RNAP inhibition in archaea. The Acidianus two-tailed virus encodes the RIP factor that binds inside the DNA-binding channel of RNAP, inhibiting transcription by occlusion of binding sites for nucleic acid and the transcription initiation factor TFB. Infection with the Sulfolobus Turreted Icosahedral Virus induces the expression of the host factor TFS4, which binds in the RNAP funnel similarly to eukaryotic transcript cleavage factors. However, TFS4 allosterically induces a widening of the DNA-binding channel which disrupts trigger loop and bridge helix motifs. Importantly, the conformational changes induced by TFS4 are closely related to inactivated states of RNAP in other domains of life indicating a deep evolutionary conservation of allosteric RNAP inhibition. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13034.map.gz emd_13034.map.gz | 128.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13034-v30.xml emd-13034-v30.xml emd-13034.xml emd-13034.xml | 40.6 KB 40.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13034_fsc.xml emd_13034_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_13034.png emd_13034.png | 121.8 KB | ||
| Filedesc metadata |  emd-13034.cif.gz emd-13034.cif.gz | 10.3 KB | ||
| Others |  emd_13034_additional_1.map.gz emd_13034_additional_1.map.gz emd_13034_half_map_1.map.gz emd_13034_half_map_1.map.gz emd_13034_half_map_2.map.gz emd_13034_half_map_2.map.gz | 10 MB 110.4 MB 110.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13034 http://ftp.pdbj.org/pub/emdb/structures/EMD-13034 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13034 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13034 | HTTPS FTP |
-Validation report
| Summary document |  emd_13034_validation.pdf.gz emd_13034_validation.pdf.gz | 862.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13034_full_validation.pdf.gz emd_13034_full_validation.pdf.gz | 862 KB | Display | |
| Data in XML |  emd_13034_validation.xml.gz emd_13034_validation.xml.gz | 19.5 KB | Display | |
| Data in CIF |  emd_13034_validation.cif.gz emd_13034_validation.cif.gz | 25.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13034 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13034 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13034 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13034 | HTTPS FTP |
-Related structure data
| Related structure data |  7oqyMC  7ok0C  7oq4C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13034.map.gz / Format: CCP4 / Size: 139.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13034.map.gz / Format: CCP4 / Size: 139.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map from Autosharpen, Phenix v1.15.2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.085 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
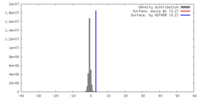
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Sharpened map from postprocess, relion v3.0. The map...
| File | emd_13034_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map from postprocess, relion v3.0. The map was obtained from Multibody refinement and it corresponds to the smaller body with higher flexibility | ||||||||||||
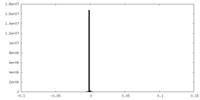
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half1 map from 3D refinement, relion v3.0
| File | emd_13034_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half1 map from 3D refinement, relion v3.0 | ||||||||||||
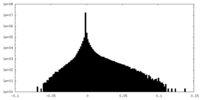
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half2 map from 3D refinement, relion v3.0
| File | emd_13034_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half2 map from 3D refinement, relion v3.0 | ||||||||||||
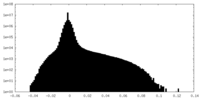
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : TFS4 bound to the RNA polymerase
+Supramolecule #1: TFS4 bound to the RNA polymerase
+Macromolecule #1: DNA-directed RNA polymerase subunit A'
+Macromolecule #2: DNA-directed RNA polymerase subunit B
+Macromolecule #3: DNA-directed RNA polymerase subunit A''
+Macromolecule #4: DNA-directed RNA polymerase subunit D
+Macromolecule #5: DNA-directed RNA polymerase subunit E
+Macromolecule #6: DNA-directed RNA polymerase, subunit F
+Macromolecule #7: DNA-directed RNA polymerase, subunit G
+Macromolecule #8: DNA-directed RNA polymerase subunit H
+Macromolecule #9: DNA-directed RNA polymerase subunit K
+Macromolecule #10: DNA-directed RNA polymerase subunit L
+Macromolecule #11: DNA-directed RNA polymerase subunit N
+Macromolecule #12: DNA-directed RNA polymerase subunit P
+Macromolecule #13: Conserved protein
+Macromolecule #14: DNA-directed RNA polymerase, subunit M (RpoM-2)
+Macromolecule #15: MAGNESIUM ION
+Macromolecule #16: ZINC ION
+Macromolecule #17: FE3-S4 CLUSTER
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
| ||||||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR Details: the grid was coated with graphene oxide after to be glow discharged and before applying the sample | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 94 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||||||||
| Details | the sample was applied twice on the grid before vetrification |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 1760 / Average exposure time: 2.0 sec. / Average electron dose: 45.2 e/Å2 Details: images were collected as movie-stacks of 40 frames in super resolution mode |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)