[English] 日本語
 Yorodumi
Yorodumi- EMDB-1237: Infectious bursal disease virus capsid assembly and maturation by... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1237 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Infectious bursal disease virus capsid assembly and maturation by structural rearrangements of a transient molecular switch. | |||||||||
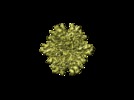
 Map data Map data | Density map of the T=1 Infectious Bursal Disease Virus (IBDV) Subviral Particle (SVP)2X2Y2Z oriented. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Infectious bursal disease virus (Gumboro virus) Infectious bursal disease virus (Gumboro virus) | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 7.2 Å | |||||||||
 Authors Authors | Luque D / Saugar I / Rodriguez JF / Verdaguer N / Garriga D / San Martin C / Velazquez-Muriel JA / Trus BL / Carrascosa JL / Caston JR | |||||||||
 Citation Citation |  Journal: J Virol / Year: 2007 Journal: J Virol / Year: 2007Title: Infectious bursal disease virus capsid assembly and maturation by structural rearrangements of a transient molecular switch. Authors: Daniel Luque / Irene Saugar / José F Rodríguez / Nuria Verdaguer / Damiá Garriga / Carmen San Martín / Javier A Velázquez-Muriel / Benes L Trus / José L Carrascosa / José R Castón /  Abstract: Infectious bursal disease virus (IBDV), a double-stranded RNA (dsRNA) virus belonging to the Birnaviridae family, is an economically important avian pathogen. The IBDV capsid is based on a single- ...Infectious bursal disease virus (IBDV), a double-stranded RNA (dsRNA) virus belonging to the Birnaviridae family, is an economically important avian pathogen. The IBDV capsid is based on a single-shelled T=13 lattice, and the only structural subunits are VP2 trimers. During capsid assembly, VP2 is synthesized as a protein precursor, called pVP2, whose 71-residue C-terminal end is proteolytically processed. The conformational flexibility of pVP2 is due to an amphipathic alpha-helix located at its C-terminal end. VP3, the other IBDV major structural protein that accomplishes numerous roles during the viral cycle, acts as a scaffolding protein required for assembly control. Here we address the molecular mechanism that defines the multimeric state of the capsid protein as hexamers or pentamers. We used a combination of three-dimensional cryo-electron microscopy maps at or close to subnanometer resolution with atomic models. Our studies suggest that the key polypeptide element, the C-terminal amphipathic alpha-helix, which acts as a transient conformational switch, is bound to the flexible VP2 C-terminal end. In addition, capsid protein oligomerization is also controlled by the progressive trimming of its C-terminal domain. The coordination of these molecular events correlates viral capsid assembly with different conformations of the amphipathic alpha-helix in the precursor capsid, as a five-alpha-helix bundle at the pentamers or an open star-like conformation at the hexamers. These results, reminiscent of the assembly pathway of positive single-stranded RNA viruses, such as nodavirus and tetravirus, add new insights into the evolutionary relationships of dsRNA viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1237.map.gz emd_1237.map.gz | 46.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1237-v30.xml emd-1237-v30.xml emd-1237.xml emd-1237.xml | 10.3 KB 10.3 KB | Display Display |  EMDB header EMDB header |
| Images |  1237.gif 1237.gif | 10.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1237 http://ftp.pdbj.org/pub/emdb/structures/EMD-1237 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1237 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1237 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1237.map.gz / Format: CCP4 / Size: 53.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1237.map.gz / Format: CCP4 / Size: 53.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density map of the T=1 Infectious Bursal Disease Virus (IBDV) Subviral Particle (SVP)2X2Y2Z oriented. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
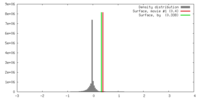
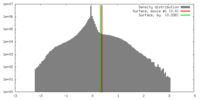
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Infectious Bursal Disease Virus Subviral Particle
| Entire | Name: Infectious Bursal Disease Virus Subviral Particle |
|---|---|
| Components |
|
-Supramolecule #1000: Infectious Bursal Disease Virus Subviral Particle
| Supramolecule | Name: Infectious Bursal Disease Virus Subviral Particle / type: sample / ID: 1000 / Oligomeric state: 60 copies of VP2 arranged in 20 trimers / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 2.8 MDa |
-Supramolecule #1: Infectious bursal disease virus
| Supramolecule | Name: Infectious bursal disease virus / type: virus / ID: 1 / Name.synonym: Gumboro virus, subviral particle / Details: IBDV Capsid Protein / NCBI-ID: 10995 / Sci species name: Infectious bursal disease virus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes / Syn species name: Gumboro virus, subviral particle |
|---|---|
| Host (natural) | Organism:  |
| Molecular weight | Experimental: 2.880 MDa |
| Virus shell | Shell ID: 1 / Name: IBDV Subviral particle / Diameter: 260 Å / T number (triangulation number): 1 |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.2 / Details: 25 mM PIPES, 150 mM NaCl, and 20 mM CaCl2 |
|---|---|
| Staining | Type: NEGATIVE Details: Samples containing were applied to one side of a holey carbon film, washed twice on water drops, blotted, and plunged into a liquid ethane bath following standard procedures |
| Grid | Details: 300 mesh holey carbon film |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 23 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 7.2 Å / Resolution method: FSC 0.33 CUT-OFF / Software - Name: em3dr2 Details: Resolution for SVP was also evaluated by FSC calculated between the full-dataset map and the SVP atomic map and for a correlation limit of 0.3, this was at 6.6 A. Number images used: 23754 |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name: SITUS and URO |
| Details | Protocol: Rigid body. Refinement was done in both Real and Fourier Sapce |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: CC and R-factor |
-Atomic model buiding 2
| Initial model | PDB ID: |
|---|---|
| Software | Name: SITUS and URO |
| Details | Protocol: Rigid body. Refinement was done in both Real and Fourier Sapce |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: CC and R-factor |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)