+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12165 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Integrator cleavage module with INTS4/9/11 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nuclease / Integrator / 3'-end processing / NUCLEAR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsnRNA 3'-end processing / snRNA processing / INTAC complex / regulation of transcription elongation by RNA polymerase II / integrator complex / Hydrolases; Acting on ester bonds; Endoribonucleases producing 3'-phosphomonoesters / RNA polymerase II transcription initiation surveillance / RNA polymerase II transcribes snRNA genes / RNA endonuclease activity / negative regulation of transforming growth factor beta receptor signaling pathway ...snRNA 3'-end processing / snRNA processing / INTAC complex / regulation of transcription elongation by RNA polymerase II / integrator complex / Hydrolases; Acting on ester bonds; Endoribonucleases producing 3'-phosphomonoesters / RNA polymerase II transcription initiation surveillance / RNA polymerase II transcribes snRNA genes / RNA endonuclease activity / negative regulation of transforming growth factor beta receptor signaling pathway / blood microparticle / protein-macromolecule adaptor activity / nucleolus / nucleoplasm / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.56 Å | |||||||||
 Authors Authors | Pfleiderer MM / Galej WP | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2021 Journal: Mol Cell / Year: 2021Title: Structure of the catalytic core of the Integrator complex. Authors: Moritz M Pfleiderer / Wojciech P Galej /  Abstract: The Integrator is a specialized 3' end-processing complex involved in cleavage and transcription termination of a subset of nascent RNA polymerase II transcripts, including small nuclear RNAs (snRNAs) ...The Integrator is a specialized 3' end-processing complex involved in cleavage and transcription termination of a subset of nascent RNA polymerase II transcripts, including small nuclear RNAs (snRNAs). We provide evidence of the modular nature of the Integrator complex by biochemically characterizing its two subcomplexes, INTS5/8 and INTS10/13/14. Using cryoelectron microscopy (cryo-EM), we determined a 3.5-Å-resolution structure of the INTS4/9/11 ternary complex, which constitutes Integrator's catalytic core. Our structure reveals the spatial organization of the catalytic nuclease INTS11, bound to its catalytically impaired homolog INTS9 via several interdependent interfaces. INTS4, a helical repeat protein, plays a key role in stabilizing nuclease domains and other components. In this assembly, all three proteins form a composite electropositive groove, suggesting a putative RNA binding path within the complex. Comparison with other 3' end-processing machineries points to distinct features and a unique architecture of the Integrator's catalytic module. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12165.map.gz emd_12165.map.gz | 150.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12165-v30.xml emd-12165-v30.xml emd-12165.xml emd-12165.xml | 19.9 KB 19.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12165_fsc.xml emd_12165_fsc.xml | 12.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_12165.png emd_12165.png | 128.6 KB | ||
| Masks |  emd_12165_msk_1.map emd_12165_msk_1.map | 160.8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-12165.cif.gz emd-12165.cif.gz | 6.9 KB | ||
| Others |  emd_12165_half_map_1.map.gz emd_12165_half_map_1.map.gz emd_12165_half_map_2.map.gz emd_12165_half_map_2.map.gz | 127.6 MB 127.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12165 http://ftp.pdbj.org/pub/emdb/structures/EMD-12165 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12165 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12165 | HTTPS FTP |
-Related structure data
| Related structure data |  7bfpMC  7bfqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12165.map.gz / Format: CCP4 / Size: 160.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12165.map.gz / Format: CCP4 / Size: 160.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
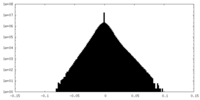
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12165_msk_1.map emd_12165_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_12165_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_12165_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Heterotrimeric cleavage module of the Integrator complex
| Entire | Name: Heterotrimeric cleavage module of the Integrator complex |
|---|---|
| Components |
|
-Supramolecule #1: Heterotrimeric cleavage module of the Integrator complex
| Supramolecule | Name: Heterotrimeric cleavage module of the Integrator complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 257 kDa/nm |
-Macromolecule #1: Integrator complex subunit 9
| Macromolecule | Name: Integrator complex subunit 9 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 73.891219 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MKLYCLSGHP TLPCNVLKFK STTIMLDCGL DMTSTLNFLP LPLVQSPRLS NLPGWSLKDG NAFLDKELKE CSGHVFVDSV PEFCLPETE LIDLSTVDVI LISNYHCMMA LPYITEHTGF TGTVYATEPT VQIGRLLMEE LVNFIERVPK AQSASLWKNK D IQRLLPSP ...String: MKLYCLSGHP TLPCNVLKFK STTIMLDCGL DMTSTLNFLP LPLVQSPRLS NLPGWSLKDG NAFLDKELKE CSGHVFVDSV PEFCLPETE LIDLSTVDVI LISNYHCMMA LPYITEHTGF TGTVYATEPT VQIGRLLMEE LVNFIERVPK AQSASLWKNK D IQRLLPSP LKDAVEVSTW RRCYTMQEVN SALSKIQLVG YSQKIELFGA VQVTPLSSGY ALGSSNWIIQ SHYEKVSYVS GS SLLTTHP QPMDQASLKN SDVLVLTGLT QIPTANPDGM VGEFCSNLAL TVRNGGNVLV PCYPSGVIYD LLECLYQYID SAG LSSVPL YFISPVANSS LEFSQIFAEW LCHNKQSKVY LPEPPFPHAE LIQTNKLKHY PSIHGDFSND FRQPCVVFTG HPSL RFGDV VHFMELWGKS SLNTVIFTEP DFSYLEALAP YQPLAMKCIY CPIDTRLNFI QVSKLLKEVQ PLHVVCPEQY TQPPP AQSH RMDLMIDCQP PAMSYRRAEV LALPFKRRYE KIEIMPELAD SLVPMEIKPG ISLATVSAVL HTKDNKHLLQ PPPRPA QPT SGKKRKRVSD DVPDCKVLKP LLSGSIPVEQ FVQTLEKHGF SDIKVEDTAK GHIVLLQEAE TLIQIEEDST HIICDND EM LRVRLRDLVL KFLQKF UniProtKB: Integrator complex subunit 9 |
-Macromolecule #2: Integrator complex subunit 4
| Macromolecule | Name: Integrator complex subunit 4 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 110.164773 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MHHHHHHHHP PSGADPMAAH LKKRVYEEFT KVVQPQEEIA TKKLRLTKPS KSAALHIDLC KATSPADALQ YLLQFARKPV EAESVEGVV RILLEHYYKE NDPSVRLKIA SLLGLLSKTA GFSPDCIMDD AINILQNEKS HQVLAQLLDT LLAIGTKLPE N QAIQMRLV ...String: MHHHHHHHHP PSGADPMAAH LKKRVYEEFT KVVQPQEEIA TKKLRLTKPS KSAALHIDLC KATSPADALQ YLLQFARKPV EAESVEGVV RILLEHYYKE NDPSVRLKIA SLLGLLSKTA GFSPDCIMDD AINILQNEKS HQVLAQLLDT LLAIGTKLPE N QAIQMRLV DVACKHLTDT SHGVRNKCLQ LLGNLGSLEK SVTKDAEGLA ARDVQKIIGD YFSDQDPRVR TAAIKAMLQL HE RGLKLHQ TIYNQACKLL SDDYEQVRSA AVQLIWVVSQ LYPESIVPIP SSNEEIRLVD DAFGKICHMV SDGSWVVRVQ AAK LLGSME QVSSHFLEQT LDKKLMSDLR RKRTAHERAK ELYSSGEFSS GRKWGDDAPK EEVDTGAVNL IESGACGAFV HGLE DEMYE VRIAAVEALC MLAQSSPSFA EKCLDFLVDM FNDEIEEVRL QSIHTMRKIS NNITLREDQL DTVLAVLEDS SRDIR EALH ELLCCTNVST KEGIHLALVE LLKNLTKYPT DRDSIWKCLK FLGSRHPTLV LPLVPELLST HPFFDTAEPD MDDPAY IAV LVLIFNAAKT CPTMPALFSD HTFRHYAYLR DSLSHLVPAL RLPGRKLVSS AVSPSIIPQE DPSQQFLQQS LERVYSL QH LDPQGAQELL EFTIRDLQRL GELQSELAGV ADFSATYLRC QLLLIKALQE KLWNVAAPLY LKQSDLASAA AKQIMEET Y KMEFMYSGVE NKQVVIIHHM RLQAKALQLI VTARTTRGLD PLFGMCEKFL QEVDFFQRYF IADLPHLQDS FVDKLLDLM PRLMTSKPAE VVKILQTMLR QSAFLHLPLP EQIHKASATI IEPAGESDNP LRFTSGLVVA LDVDATLEHV QDPQNTVKVQ VLYPDGQAQ MIHPKPADFR NPGPGRHRLI TQVYLSHTAW TEACQVEVRL LLAYNSSARI PKCPWMEGGE MSPQVETSIE G TIPFSKPV KVYIMPKPAR R UniProtKB: Integrator complex subunit 4 |
-Macromolecule #3: Integrator complex subunit 11
| Macromolecule | Name: Integrator complex subunit 11 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on ester bonds; Endoribonucleases producing 3'-phosphomonoesters |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 72.690008 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MDEKTTGWRG GHVVEGLAGE LEQLRARLEH HPQGQREPPP SGADPMPEIR VTPLGAGQDV GRSCILVSIA GKNVMLDCGM HMGFNDDRR FPDFSYITQN GRLTDFLDCV IISHFHLDHC GALPYFSEMV GYDGPIYMTH PTQAICPILL EDYRKIAVDK K GEANFFTS ...String: MDEKTTGWRG GHVVEGLAGE LEQLRARLEH HPQGQREPPP SGADPMPEIR VTPLGAGQDV GRSCILVSIA GKNVMLDCGM HMGFNDDRR FPDFSYITQN GRLTDFLDCV IISHFHLDHC GALPYFSEMV GYDGPIYMTH PTQAICPILL EDYRKIAVDK K GEANFFTS QMIKDCMKKV VAVHLHQTVQ VDDELEIKAY YAGHVLGAAM FQIKVGSESV VYTGDYNMTP DRHLGAAWID KC RPNLLIT ESTYATTIRD SKRCRERDFL KKVHETVERG GKVLIPVFAL GRAQELCILL ETFWERMNLK VPIYFSTGLT EKA NHYYKL FIPWTNQKIR KTFVQRNMFE FKHIKAFDRA FADNPGPMVV FATPGMLHAG QSLQIFRKWA GNEKNMVIMP GYCV QGTVG HKILSGQRKL EMEGRQVLEV KMQVEYMSFS AHADAKGIMQ LVGQAEPESV LLVHGEAKKM EFLKQKIEQE LRVNC YMPA NGETVTLLTS PSIPVGISLG LLKREMAQGL LPEAKKPRLL HGTLIMKDSN FRLVSSEQAL KELGLAEHQL RFTCRV HLH DTRKEQETAL RVYSHLKSVL KDHCVQHLPD GSVTVESVLL QAAAPSEDPG TKVLLVSWTY QDEELGSFLT SLLKKGL PQ APS UniProtKB: Integrator complex subunit 11 |
-Macromolecule #4: Unknown
| Macromolecule | Name: Unknown / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 3.08179 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.8 / Details: 150 mM KCl, 20 mM HEPES-KOH pH 7.8 |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Number grids imaged: 2 / Number real images: 19268 / Average exposure time: 5.0 sec. / Average electron dose: 44.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 0.5 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 165000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-7bfp: |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)