[English] 日本語
 Yorodumi
Yorodumi- EMDB-12015: Programmable icosahedral shell system for virus trapping: DNA-ori... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12015 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Programmable icosahedral shell system for virus trapping: DNA-origami T=9 triangle (hexamer 2) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species | synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 15.0 Å | |||||||||
 Authors Authors | Sigl C / Willner EM / Engelen W / Kretzmann JA / Sachenbacher K / Liedl A / Kolbe F / Wilsch F / Aghvami SA / Protzer U ...Sigl C / Willner EM / Engelen W / Kretzmann JA / Sachenbacher K / Liedl A / Kolbe F / Wilsch F / Aghvami SA / Protzer U / Hagan MF / Fraden S / Dietz H | |||||||||
| Funding support |  Germany, European Union, 2 items Germany, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Mater / Year: 2021 Journal: Nat Mater / Year: 2021Title: Programmable icosahedral shell system for virus trapping. Authors: Christian Sigl / Elena M Willner / Wouter Engelen / Jessica A Kretzmann / Ken Sachenbacher / Anna Liedl / Fenna Kolbe / Florian Wilsch / S Ali Aghvami / Ulrike Protzer / Michael F Hagan / ...Authors: Christian Sigl / Elena M Willner / Wouter Engelen / Jessica A Kretzmann / Ken Sachenbacher / Anna Liedl / Fenna Kolbe / Florian Wilsch / S Ali Aghvami / Ulrike Protzer / Michael F Hagan / Seth Fraden / Hendrik Dietz /   Abstract: Broad-spectrum antiviral platforms that can decrease or inhibit viral infection would alleviate many threats to global public health. Nonetheless, effective technologies of this kind are still not ...Broad-spectrum antiviral platforms that can decrease or inhibit viral infection would alleviate many threats to global public health. Nonetheless, effective technologies of this kind are still not available. Here, we describe a programmable icosahedral canvas for the self-assembly of icosahedral shells that have viral trapping and antiviral properties. Programmable triangular building blocks constructed from DNA assemble with high yield into various shell objects with user-defined geometries and apertures. We have created shells with molecular masses ranging from 43 to 925 MDa (8 to 180 subunits) and with internal cavity diameters of up to 280 nm. The shell interior can be functionalized with virus-specific moieties in a modular fashion. We demonstrate this virus-trapping concept by engulfing hepatitis B virus core particles and adeno-associated viruses. We demonstrate the inhibition of hepatitis B virus core interactions with surfaces in vitro and the neutralization of infectious adeno-associated viruses exposed to human cells. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12015.map.gz emd_12015.map.gz | 349.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12015-v30.xml emd-12015-v30.xml emd-12015.xml emd-12015.xml | 11.5 KB 11.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12015_fsc.xml emd_12015_fsc.xml | 16.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_12015.png emd_12015.png | 55.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12015 http://ftp.pdbj.org/pub/emdb/structures/EMD-12015 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12015 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12015 | HTTPS FTP |
-Validation report
| Summary document |  emd_12015_validation.pdf.gz emd_12015_validation.pdf.gz | 266.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12015_full_validation.pdf.gz emd_12015_full_validation.pdf.gz | 265.7 KB | Display | |
| Data in XML |  emd_12015_validation.xml.gz emd_12015_validation.xml.gz | 15 KB | Display | |
| Data in CIF |  emd_12015_validation.cif.gz emd_12015_validation.cif.gz | 20.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12015 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12015 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12015 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12015 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12015.map.gz / Format: CCP4 / Size: 376.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12015.map.gz / Format: CCP4 / Size: 376.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.79 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
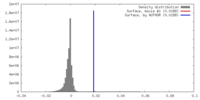
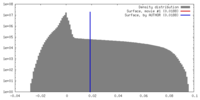
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : DNA-origami T=9 triangle (hexamer 2)
| Entire | Name: DNA-origami T=9 triangle (hexamer 2) |
|---|---|
| Components |
|
-Supramolecule #1: DNA-origami T=9 triangle (hexamer 2)
| Supramolecule | Name: DNA-origami T=9 triangle (hexamer 2) / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Recombinant expression | Organism: synthetic construct (others) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)