[English] 日本語
 Yorodumi
Yorodumi- EMDB-11621: Class 4 of three-dimensional classification of single asymmetric ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11621 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Class 4 of three-dimensional classification of single asymmetric units of RV-A89 into six classes. The class averages reveal differences at the RNA/protein interfaces. | |||||||||
 Map data Map data | Class 4 of three-dimensional classification of single asymmetric units of RV-A89 into six classes. The class averages reveal differences at the RNA/protein interfaces. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Rhinovirus A Rhinovirus A | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Blaas D | |||||||||
| Funding support |  Austria, 2 items Austria, 2 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2020 Journal: Commun Biol / Year: 2020Title: Individual subunits of a rhinovirus causing common cold exhibit largely different protein-RNA contact site conformations. Authors: Dieter Blaas /  Abstract: Rhinoviruses cause the common cold. They are icosahedral, built from sixty copies each of the capsid proteins VP1 through VP4 arranged in a pseudo T = 3 lattice. This shell encases a ss(+) RNA ...Rhinoviruses cause the common cold. They are icosahedral, built from sixty copies each of the capsid proteins VP1 through VP4 arranged in a pseudo T = 3 lattice. This shell encases a ss(+) RNA genome. Three-D classification of single and oligomeric asymmetric units computationally excised from a 2.9 Å cryo-EM density map of rhinovirus A89, showed that VP4 and the N-terminal extension of VP1 adopt different conformations within the otherwise identical 3D-structures. Analysis of up to sixty classes of single subunits and of six classes of subunit dimers, trimers, and pentamers revealed different orientations of the amino acid residues at the interface with the RNA suggesting that local asymmetry is dictated by disparities of the interacting nucleotide sequences. The different conformations escape detection by 3-D structure determination of entire virions with the conformational heterogeneity being only indicated by low density. My results do not exclude that the RNA follows a conserved assembly mechanism, contacting most or all asymmetric units in a specific way. However, as suggested by the gradual loss of asymmetry with increasing oligomerization and the 3D-structure of entire virions reconstructed by using Euler angles selected in the classification of single subunits, RNA path and/or folding likely differ from virion to virion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11621.map.gz emd_11621.map.gz | 3.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11621-v30.xml emd-11621-v30.xml emd-11621.xml emd-11621.xml | 8.1 KB 8.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_11621.png emd_11621.png | 207.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11621 http://ftp.pdbj.org/pub/emdb/structures/EMD-11621 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11621 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11621 | HTTPS FTP |
-Validation report
| Summary document |  emd_11621_validation.pdf.gz emd_11621_validation.pdf.gz | 196.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11621_full_validation.pdf.gz emd_11621_full_validation.pdf.gz | 195.9 KB | Display | |
| Data in XML |  emd_11621_validation.xml.gz emd_11621_validation.xml.gz | 7.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11621 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11621 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11621 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11621 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11621.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11621.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Class 4 of three-dimensional classification of single asymmetric units of RV-A89 into six classes. The class averages reveal differences at the RNA/protein interfaces. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.97 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
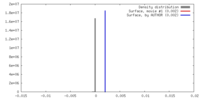
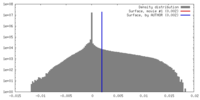
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Rhinovirus A
| Entire | Name:  Rhinovirus A Rhinovirus A |
|---|---|
| Components |
|
-Supramolecule #1: Rhinovirus A
| Supramolecule | Name: Rhinovirus A / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 147711 / Sci species name: Rhinovirus A / Sci species strain: Rhinovirus A89 / Virus type: VIRION / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Virus shell | Shell ID: 1 / Diameter: 302.0 Å |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 107054 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)