[English] 日本語
 Yorodumi
Yorodumi- EMDB-1061: Inositol 1,4,5-trisphosphate receptor contains multiple cavities ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1061 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Inositol 1,4,5-trisphosphate receptor contains multiple cavities and L-shaped ligand-binding domains. | |||||||||
 Map data Map data | 3D volume data | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 15.0 Å | |||||||||
 Authors Authors | Sato C / Hamada K / Ogura T / Miyazawa A / Iwasaki K / Hiroaki Y / Tani K / Terauchi A / Fujiyoshi Y / Mikoshiba K | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2004 Journal: J Mol Biol / Year: 2004Title: Inositol 1,4,5-trisphosphate receptor contains multiple cavities and L-shaped ligand-binding domains. Authors: Chikara Sato / Kozo Hamada / Toshihiko Ogura / Atsuo Miyazawa / Kenji Iwasaki / Yoko Hiroaki / Kazutoshi Tani / Akiko Terauchi / Yoshinori Fujiyoshi / Katsuhiko Mikoshiba /  Abstract: Calcium concentrations are strictly regulated in all biological cells, and one of the key molecules responsible for this regulation is the inositol 1,4,5-trisphosphate receptor, which was known to ...Calcium concentrations are strictly regulated in all biological cells, and one of the key molecules responsible for this regulation is the inositol 1,4,5-trisphosphate receptor, which was known to form a homotetrameric Ca(2+) channel in the endoplasmic reticulum. The receptor is involved in neuronal transmission via Ca(2+) signaling and for many other functions that relate to morphological and physiological processes in living organisms. We analysed the three-dimensional structure of the ligand-free form of the receptor based on a single-particle technique using an originally developed electron microscope equipped with a helium-cooled specimen stage and an automatic particle picking system. We propose a model that explains the complex mechanism for the regulation of Ca(2+) release by co-agonists, Ca(2+), inositol 1,4,5-trisphosphate based on the structure of multiple internal cavities and a porous balloon-shaped cytoplasmic domain containing a prominent L-shaped density which was assigned by the X-ray structure of the inositol 1,4,5-trisphosphate binding domain. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1061.map.gz emd_1061.map.gz | 3.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1061-v30.xml emd-1061-v30.xml emd-1061.xml emd-1061.xml | 7.1 KB 7.1 KB | Display Display |  EMDB header EMDB header |
| Images |  1061.gif 1061.gif | 24 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1061 http://ftp.pdbj.org/pub/emdb/structures/EMD-1061 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1061 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1061 | HTTPS FTP |
-Validation report
| Summary document |  emd_1061_validation.pdf.gz emd_1061_validation.pdf.gz | 225.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1061_full_validation.pdf.gz emd_1061_full_validation.pdf.gz | 224.9 KB | Display | |
| Data in XML |  emd_1061_validation.xml.gz emd_1061_validation.xml.gz | 4.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1061 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1061 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1061 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1061 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1061.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1061.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D volume data | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.6 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
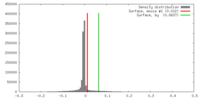
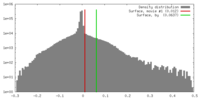
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : IP3 receptor from mouse
| Entire | Name: IP3 receptor from mouse |
|---|---|
| Components |
|
-Supramolecule #1000: IP3 receptor from mouse
| Supramolecule | Name: IP3 receptor from mouse / type: sample / ID: 1000 / Oligomeric state: tetramer / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 1.3 MDa |
-Macromolecule #1: IP3 receptor
| Macromolecule | Name: IP3 receptor / type: protein_or_peptide / ID: 1 / Name.synonym: IP3R / Details: in 1mM EGTA / Number of copies: 4 / Oligomeric state: tetramer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 1.3 MDa |
| Recombinant expression | Organism: natural source (unknown) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE |
|---|
- Electron microscopy
Electron microscopy
| Microscope | JEOL KYOTO-3000SFF |
|---|---|
| Temperature | Average: 4.2 K |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder: top entry / Specimen holder model: OTHER |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C4 (4 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 15.0 Å / Resolution method: FSC 3 SIGMA CUT-OFF |
|---|
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)