+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0945 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | High resolution architecture of curli complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | curli / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcurli secretion complex / curli assembly / protein secretion by the type VIII secretion system / protein transmembrane transport / single-species biofilm formation / cell outer membrane / outer membrane-bounded periplasmic space / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
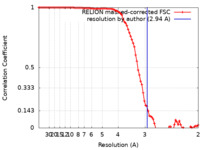
| Method | single particle reconstruction / cryo EM / Resolution: 2.94 Å | |||||||||
 Authors Authors | Zhang M / Shi H | |||||||||
 Citation Citation |  Journal: PLoS Biol / Year: 2020 Journal: PLoS Biol / Year: 2020Title: Cryo-EM structure of the nonameric CsgG-CsgF complex and its implications for controlling curli biogenesis in Enterobacteriaceae. Authors: Manfeng Zhang / Huigang Shi / Xuemei Zhang / Xinzheng Zhang / Yihua Huang /  Abstract: Curli play critical roles in biofilm formation, host cell adhesion, and colonization of inert surfaces in many Enterobacteriaceae. In Escherichia coli, curli biogenesis requires 7 curli-specific gene ...Curli play critical roles in biofilm formation, host cell adhesion, and colonization of inert surfaces in many Enterobacteriaceae. In Escherichia coli, curli biogenesis requires 7 curli-specific gene (csg) products-CsgA through G-working in concert. Of them, CsgG and CsgF are 2 outer membrane (OM)-localized components that consists of the core apparatus for secretion and assembly of curli structural subunits, CsgB and CsgA. Here, we report the cryogenic electron microscopy (cryo-EM) structure of CsgG in complex with CsgF from E. coli. The structure reveals that CsgF forms a stable complex with CsgG via a 1:1 stoichiometry by lining the upper lumen of the nonameric CsgG channel via its N-terminal 27 residues, forming a funnel-like entity plugged in the CsgG channel and creating a unique secretion channel with 2 constriction regions, consistent with the recently reported structure of the CsgG-CsgF complex. Functional studies indicate that export of CsgF to the cell surface requires the CsgG channel, and CsgF not only functions as an adaptor that bridges CsgB with CsgG but also may play important roles in controlling the rates of translocation and/or polymerization for curli structural subunits. Importantly, we found that a series of CsgF-derived peptides are able to efficiently inhibit curli production to E. coli when administrated exogenously, highlighting a potential strategy to interfere biofilm formation in E. coli strains. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
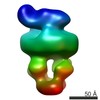
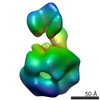
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0945.map.gz emd_0945.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0945-v30.xml emd-0945-v30.xml emd-0945.xml emd-0945.xml | 20.3 KB 20.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0945_fsc.xml emd_0945_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_0945.png emd_0945.png | 60.8 KB | ||
| Filedesc metadata |  emd-0945.cif.gz emd-0945.cif.gz | 6.1 KB | ||
| Others |  emd_0945_half_map_1.map.gz emd_0945_half_map_1.map.gz emd_0945_half_map_2.map.gz emd_0945_half_map_2.map.gz | 49.1 MB 49 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0945 http://ftp.pdbj.org/pub/emdb/structures/EMD-0945 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0945 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0945 | HTTPS FTP |
-Related structure data
| Related structure data |  6lqhMC  0947C  6lqjC  7brmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0945.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0945.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
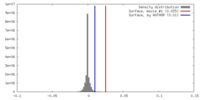
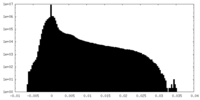
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #2
| File | emd_0945_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
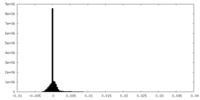
| Density Histograms |
-Half map: #1
| File | emd_0945_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
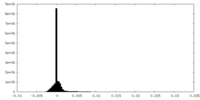
| Density Histograms |
- Sample components
Sample components
-Entire : curli core complex of CsgG and CsgF
| Entire | Name: curli core complex of CsgG and CsgF |
|---|---|
| Components |
|
-Supramolecule #1: curli core complex of CsgG and CsgF
| Supramolecule | Name: curli core complex of CsgG and CsgF / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Curli production assembly/transport component CsgG
| Macromolecule | Name: Curli production assembly/transport component CsgG / type: protein_or_peptide / ID: 1 / Number of copies: 9 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 31.62618 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQRLFLLVAV MLLSGCLTAP PKEAARPTLM PRAQSYKDLT HLPAPTGKIF VSVYNIQDET GQFKPYPASN FSTAVPQSAT AMLVTALKD SRWFIPLERQ GLQNLLNERK IIRAAQENGT VAINNRIPLQ SLTAANIMVE GSIIGYESNV KSGGVGARYF G IGADTQYQ ...String: MQRLFLLVAV MLLSGCLTAP PKEAARPTLM PRAQSYKDLT HLPAPTGKIF VSVYNIQDET GQFKPYPASN FSTAVPQSAT AMLVTALKD SRWFIPLERQ GLQNLLNERK IIRAAQENGT VAINNRIPLQ SLTAANIMVE GSIIGYESNV KSGGVGARYF G IGADTQYQ LDQIAVNLRV VNVSTGEILS SVNTSKTILS YEVQAGVFRF IDYQRLLEGE VGYTSNEPVM LCLMSAIETG VI FLINDGI DRGLWDLQNK AERQNDILVK YRHMSVPPES WSHPQFEK UniProtKB: Curli production assembly/transport component CsgG |
-Macromolecule #2: Curli production assembly/transport component CsgF
| Macromolecule | Name: Curli production assembly/transport component CsgF / type: protein_or_peptide / ID: 2 / Number of copies: 9 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.894586 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRVKHAVVLL MLISPLSWAG TMTFQFRNPN FGGNPNNGAF LLNSAQAQNS YKDPSYNDDF GIETPSALDN FTQAIQSQIL GGLLSNINT GKPGRMVTND YIVDIANRDG QLQLNVTDRK TGQTSTIQVS GLQNNSTDFH HHHHH UniProtKB: Curli production assembly/transport component CsgF |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 298 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Temperature | Min: 70.0 K |
| Alignment procedure | Coma free - Residual tilt: 0.1 mrad |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 BASE (4k x 4k) / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Number grids imaged: 1 / Number real images: 2500 / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus min: 1.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)