+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0312 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
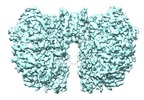
| Title | Structure of McrBC without DNA binding domains (Class 2) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AAA+ superfamily / restriction enzyme / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationtype IV site-specific deoxyribonuclease activity / restriction endodeoxyribonuclease activity / endonuclease complex / double-stranded methylated DNA binding / hemi-methylated DNA-binding / Hydrolases; Acting on ester bonds; Endodeoxyribonucleases producing 5'-phosphomonoesters / DNA catabolic process / DNA restriction-modification system / endonuclease activity / GTPase activity ...type IV site-specific deoxyribonuclease activity / restriction endodeoxyribonuclease activity / endonuclease complex / double-stranded methylated DNA binding / hemi-methylated DNA-binding / Hydrolases; Acting on ester bonds; Endodeoxyribonucleases producing 5'-phosphomonoesters / DNA catabolic process / DNA restriction-modification system / endonuclease activity / GTPase activity / GTP binding / ATP hydrolysis activity / DNA binding / ATP binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |   | |||||||||
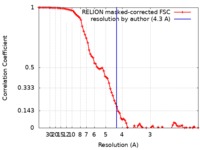
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | |||||||||
 Authors Authors | Itoh Y / Nirwan N | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Structure-based mechanism for activation of the AAA+ GTPase McrB by the endonuclease McrC. Authors: Neha Nirwan / Yuzuru Itoh / Pratima Singh / Sutirtha Bandyopadhyay / Kutti R Vinothkumar / Alexey Amunts / Kayarat Saikrishnan /    Abstract: The AAA+ GTPase McrB powers DNA cleavage by the endonuclease McrC. The GTPase itself is activated by McrC. The architecture of the GTPase and nuclease complex, and the mechanism of their activation ...The AAA+ GTPase McrB powers DNA cleavage by the endonuclease McrC. The GTPase itself is activated by McrC. The architecture of the GTPase and nuclease complex, and the mechanism of their activation remained unknown. Here, we report a 3.6 Å structure of a GTPase-active and DNA-binding deficient construct of McrBC. Two hexameric rings of McrB are bridged by McrC dimer. McrC interacts asymmetrically with McrB protomers and inserts a stalk into the pore of the ring, reminiscent of the γ subunit complexed to αβ of F-ATPase. Activation of the GTPase involves conformational changes of residues essential for hydrolysis. Three consecutive nucleotide-binding pockets are occupied by the GTP analogue 5'-guanylyl imidodiphosphate and the next three by GDP, which is suggestive of sequential GTP hydrolysis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0312.map.gz emd_0312.map.gz | 7.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0312-v30.xml emd-0312-v30.xml emd-0312.xml emd-0312.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0312_fsc.xml emd_0312_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_0312.png emd_0312.png | 302.9 KB | ||
| Masks |  emd_0312_msk_1.map emd_0312_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-0312.cif.gz emd-0312.cif.gz | 6.7 KB | ||
| Others |  emd_0312_half_map_1.map.gz emd_0312_half_map_1.map.gz emd_0312_half_map_2.map.gz emd_0312_half_map_2.map.gz | 39.7 MB 39.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0312 http://ftp.pdbj.org/pub/emdb/structures/EMD-0312 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0312 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0312 | HTTPS FTP |
-Related structure data
| Related structure data |  6hz6MC  0310C  0311C  0313C  0314C  0315C  6hz4C  6hz5C  6hz7C  6hz8C  6hz9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0312.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0312.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
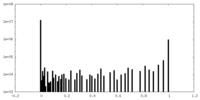
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
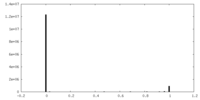
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_0312_msk_1.map emd_0312_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
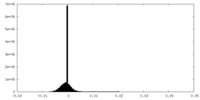
| Density Histograms |
-Half map: #2
| File | emd_0312_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
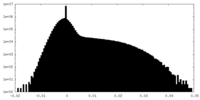
| Density Histograms |
-Half map: #1
| File | emd_0312_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : McrB and McrC complex without DNA binding domains
| Entire | Name: McrB and McrC complex without DNA binding domains |
|---|---|
| Components |
|
-Supramolecule #1: McrB and McrC complex without DNA binding domains
| Supramolecule | Name: McrB and McrC complex without DNA binding domains / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: The N-terminal DNA binding domain of McrB is truncated |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 510 KDa |
-Macromolecule #1: 5-methylcytosine-specific restriction enzyme B
| Macromolecule | Name: 5-methylcytosine-specific restriction enzyme B / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO EC number: Hydrolases; Acting on ester bonds; Endodeoxyribonucleases producing 5'-phosphomonoesters |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 35.758492 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSKTESYCLE DALNDLFIPE TTIETILKRL TIKKNIILQG PPGVGKTFVA RRLAYLLTGE KAPQRVNMVQ FHQSYSYEDF IQGYRPNGV GFRRKDGIFY NFCQQAKEQP EKKYIFIIDE INRANLSKVF GEVMMLMEHD KRGENWSVPL TYSENDEERF Y VPENVYII ...String: MSKTESYCLE DALNDLFIPE TTIETILKRL TIKKNIILQG PPGVGKTFVA RRLAYLLTGE KAPQRVNMVQ FHQSYSYEDF IQGYRPNGV GFRRKDGIFY NFCQQAKEQP EKKYIFIIDE INRANLSKVF GEVMMLMEHD KRGENWSVPL TYSENDEERF Y VPENVYII GLMNTADRSL AVVDYALRRR FSFIDIEPGF DTPQFRNFLL NKKAEPSFVE SLCQKMNELN QEISKEATIL GK GFRIGHS YFCCGLEDGT SPDTQWLNEI VMTDIAPLLE EYFFDDPYKQ QKWTNKLLGD SSGSHHHHHH UniProtKB: Type IV methyl-directed restriction enzyme EcoKMcrB subunit |
-Macromolecule #2: Protein McrC
| Macromolecule | Name: Protein McrC / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 40.643625 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEQPVIPVRN IYYMLTYAWG YLQEIKQANL EAIPGNNLLD ILGYVLNKGV LQLSRRGLEL DYNPNTEIIP GIKGRIEFAK TIRGFHLNH GKTVSTFDML NEDTLANRII KSTLAILIKH EKLNSTIRDE ARSLYRKLPG ISTLHLTPQH FSYLNGGKNT R YYKFVISV ...String: MEQPVIPVRN IYYMLTYAWG YLQEIKQANL EAIPGNNLLD ILGYVLNKGV LQLSRRGLEL DYNPNTEIIP GIKGRIEFAK TIRGFHLNH GKTVSTFDML NEDTLANRII KSTLAILIKH EKLNSTIRDE ARSLYRKLPG ISTLHLTPQH FSYLNGGKNT R YYKFVISV CKFIVNNSIP GQNKGHYRFY DFERNEKEMS LLYQKFLYEF CRRELTSANT TRSYLKWDAS SISDQSLNLL PR METDITI RSSEKILIVD AKYYKSIFSR RMGTEKFHSQ NLYQLMNYLW SLKPENGENI GGLLIYPHVD TAVKHRYKIN GFD IGLCTV NLGQEWPCIH QELLDIFDEY LK UniProtKB: Type IV methyl-directed restriction enzyme EcoKMcrBC |
-Macromolecule #3: PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER / type: ligand / ID: 3 / Number of copies: 6 / Formula: GNP |
|---|---|
| Molecular weight | Theoretical: 522.196 Da |
| Chemical component information |  ChemComp-GNP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 6 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 6 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.0 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||||||||
| Grid | Model: C-flat-2/2 4C / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 2-20 / Number grids imaged: 1 / Number real images: 3326 / Average exposure time: 8.0 sec. / Average electron dose: 1.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 5.0 µm / Calibrated defocus min: 0.3 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 74 / Target criteria: Cross-correlation coefficient |
|---|---|
| Output model |  PDB-6hz6: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)