[English] 日本語
 Yorodumi
Yorodumi- EMDB-0289: Structure of a functional obligate respiratory supercomplex from ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0289 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a functional obligate respiratory supercomplex from Mycobacterium smegmatis | |||||||||
 Map data Map data | Respiratory supercomplex from Mycobacterium smegmatis | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Membrane Protein / Cryo-EM / Respiratory supercomplex / Mycobacterium / ELECTRON TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationaerobic electron transport chain / cytochrome-c oxidase / oxidative phosphorylation / quinol-cytochrome-c reductase / quinol-cytochrome-c reductase activity / cytochrome-c oxidase activity / electron transport coupled proton transport / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen / ATP synthesis coupled electron transport / respiratory electron transport chain ...aerobic electron transport chain / cytochrome-c oxidase / oxidative phosphorylation / quinol-cytochrome-c reductase / quinol-cytochrome-c reductase activity / cytochrome-c oxidase activity / electron transport coupled proton transport / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen / ATP synthesis coupled electron transport / respiratory electron transport chain / monooxygenase activity / electron transport chain / 2 iron, 2 sulfur cluster binding / iron ion binding / copper ion binding / heme binding / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Mycobacterium smegmatis str. MC2 155 (bacteria) / Mycobacterium smegmatis str. MC2 155 (bacteria) /  Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) / Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) /  Mycobacterium smegmatis MC2 155 (bacteria) Mycobacterium smegmatis MC2 155 (bacteria) | |||||||||
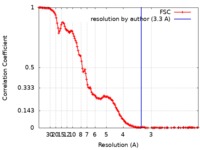
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Wiseman B / Nitharwal RG | |||||||||
| Funding support |  Canada, Canada,  Sweden, 2 items Sweden, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2018 Journal: Nat Struct Mol Biol / Year: 2018Title: Structure of a functional obligate complex IIIIV respiratory supercomplex from Mycobacterium smegmatis. Authors: Benjamin Wiseman / Ram Gopal Nitharwal / Olga Fedotovskaya / Jacob Schäfer / Hui Guo / Qie Kuang / Samir Benlekbir / Dan Sjöstrand / Pia Ädelroth / John L Rubinstein / Peter Brzezinski / Martin Högbom /    Abstract: In the mycobacterial electron-transport chain, respiratory complex III passes electrons from menaquinol to complex IV, which in turn reduces oxygen, the terminal acceptor. Electron transfer is ...In the mycobacterial electron-transport chain, respiratory complex III passes electrons from menaquinol to complex IV, which in turn reduces oxygen, the terminal acceptor. Electron transfer is coupled to transmembrane proton translocation, thus establishing the electrochemical proton gradient that drives ATP synthesis. We isolated, biochemically characterized, and determined the structure of the obligate IIIIV supercomplex from Mycobacterium smegmatis, a model for Mycobacterium tuberculosis. The supercomplex has quinol:O oxidoreductase activity without exogenous cytochrome c and includes a superoxide dismutase subunit that may detoxify reactive oxygen species produced during respiration. We found menaquinone bound in both the Q and Q sites of complex III. The complex III-intrinsic diheme cytochrome cc subunit, which functionally replaces both cytochrome c and soluble cytochrome c in canonical electron-transport chains, displays two conformations: one in which it provides a direct electronic link to complex IV and another in which it serves as an electrical switch interrupting the connection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0289.map.gz emd_0289.map.gz | 10.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0289-v30.xml emd-0289-v30.xml emd-0289.xml emd-0289.xml | 36.2 KB 36.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0289_fsc.xml emd_0289_fsc.xml | 15.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_0289.png emd_0289.png | 59.6 KB | ||
| Filedesc metadata |  emd-0289.cif.gz emd-0289.cif.gz | 10.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0289 http://ftp.pdbj.org/pub/emdb/structures/EMD-0289 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0289 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0289 | HTTPS FTP |
-Validation report
| Summary document |  emd_0289_validation.pdf.gz emd_0289_validation.pdf.gz | 405.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0289_full_validation.pdf.gz emd_0289_full_validation.pdf.gz | 405.2 KB | Display | |
| Data in XML |  emd_0289_validation.xml.gz emd_0289_validation.xml.gz | 13.7 KB | Display | |
| Data in CIF |  emd_0289_validation.cif.gz emd_0289_validation.cif.gz | 18.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0289 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0289 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0289 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0289 | HTTPS FTP |
-Related structure data
| Related structure data |  6hwhMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0289.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0289.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Respiratory supercomplex from Mycobacterium smegmatis | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Respiratory Supercomplex from Mycobacterium smegmatis
+Supramolecule #1: Respiratory Supercomplex from Mycobacterium smegmatis
+Macromolecule #1: Ubiquinol-cytochrome c reductase iron-sulfur subunit
+Macromolecule #2: Co-purified unknown transmembrane helices built as polyALA
+Macromolecule #3: Co-purified unknown transmembrane helices built as polyALA
+Macromolecule #4: Co-purified unknown peptide built as polyALA
+Macromolecule #5: Co-purified unknown peptide built as polyALA
+Macromolecule #6: Cytochrome bc1 complex cytochrome c subunit
+Macromolecule #7: Cytochrome c oxidase subunit 2
+Macromolecule #8: MSMEG_4693
+Macromolecule #9: Uncharacterized protein MSMEG_4692/MSMEI_4575
+Macromolecule #10: Cytochrome c oxidase subunit 1
+Macromolecule #11: Cytochrome c oxidase polypeptide 4
+Macromolecule #12: Cytochrome c oxidase subunit 3
+Macromolecule #13: Ubiquinol-cytochrome C reductase QcrB
+Macromolecule #14: FE2/S2 (INORGANIC) CLUSTER
+Macromolecule #15: CARDIOLIPIN
+Macromolecule #16: MENAQUINONE-9
+Macromolecule #17: COPPER (II) ION
+Macromolecule #18: HEME-AS
+Macromolecule #19: HEME C
+Macromolecule #20: PROTOPORPHYRIN IX CONTAINING FE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. / Pretreatment - Atmosphere: AIR / Details: 40 seconds at 20 mA |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 5316 / Average exposure time: 60.0 sec. / Average electron dose: 43.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Model was built de novo; except for chains i,j which are polyALA homology models rigid-body docked into low resolution density. |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER / Target criteria: Cross-correlation coefficient |
| Output model |  PDB-6hwh: |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)