[English] 日本語
 Yorodumi
Yorodumi- EMDB-0198: Synthetic Self-assembling ADDomer Platform for Highly Efficient V... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0198 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Synthetic Self-assembling ADDomer Platform for Highly Efficient Vaccination by Genetically-encoded Multi-epitope Display | |||||||||
 Map data Map data | ADDomer dodecahedron (engineered viral like particle derived from Ad3 Penton base protein). RELION Post Processed (Masked and B-factor sharpened map). | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | engineered / virus / like / particle / ADDomer / Ad3 / penton / base / adenovirus / synthetic / vaccine / biobrick / human / homosapien / dodecahedron / icosohedral / epitode / display / scaffold / VIRUS LIKE PARTICLE | |||||||||
| Function / homology | Adenovirus penton base protein / Adenovirus penton base protein / T=25 icosahedral viral capsid / endocytosis involved in viral entry into host cell / virion attachment to host cell / host cell nucleus / structural molecule activity / metal ion binding / Penton protein Function and homology information Function and homology information | |||||||||
| Biological species |  Human adenovirus B3 / Human adenovirus B3 /  Human adenovirus B serotype 3 Human adenovirus B serotype 3 | |||||||||
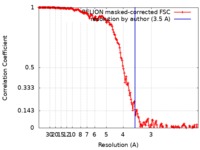
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Bufton JC / Berger I | |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2019 Journal: Sci Adv / Year: 2019Title: Synthetic self-assembling ADDomer platform for highly efficient vaccination by genetically encoded multiepitope display. Authors: Charles Vragniau / Joshua C Bufton / Frédéric Garzoni / Emilie Stermann / Fruzsina Rabi / Céline Terrat / Mélanie Guidetti / Véronique Josserand / Matt Williams / Christopher J Woods / ...Authors: Charles Vragniau / Joshua C Bufton / Frédéric Garzoni / Emilie Stermann / Fruzsina Rabi / Céline Terrat / Mélanie Guidetti / Véronique Josserand / Matt Williams / Christopher J Woods / Gerardo Viedma / Phil Bates / Bernard Verrier / Laurence Chaperot / Christiane Schaffitzel / Imre Berger / Pascal Fender /   Abstract: Self-assembling virus-like particles represent highly attractive tools for developing next-generation vaccines and protein therapeutics. We created ADDomer, an adenovirus-derived multimeric protein- ...Self-assembling virus-like particles represent highly attractive tools for developing next-generation vaccines and protein therapeutics. We created ADDomer, an adenovirus-derived multimeric protein-based self-assembling nanoparticle scaffold engineered to facilitate plug-and-play display of multiple immunogenic epitopes from pathogens. We used cryo-electron microscopy at near-atomic resolution and implemented novel, cost-effective, high-performance cloud computing to reveal architectural features in unprecedented detail. We analyzed ADDomer interaction with components of the immune system and developed a promising first-in-kind ADDomer-based vaccine candidate to combat emerging Chikungunya infectious disease, exemplifying the potential of our approach. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0198.map.gz emd_0198.map.gz | 53.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0198-v30.xml emd-0198-v30.xml emd-0198.xml emd-0198.xml | 13.5 KB 13.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0198_fsc.xml emd_0198_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_0198.png emd_0198.png | 254.4 KB | ||
| Filedesc metadata |  emd-0198.cif.gz emd-0198.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0198 http://ftp.pdbj.org/pub/emdb/structures/EMD-0198 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0198 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0198 | HTTPS FTP |
-Related structure data
| Related structure data |  6hcrMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0198.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0198.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ADDomer dodecahedron (engineered viral like particle derived from Ad3 Penton base protein). RELION Post Processed (Masked and B-factor sharpened map). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : ADDomer: An engineered viral like particle derived from penton ba...
| Entire | Name: ADDomer: An engineered viral like particle derived from penton base protein of Human adenovirus B3 (Ad3). |
|---|---|
| Components |
|
-Supramolecule #1: ADDomer: An engineered viral like particle derived from penton ba...
| Supramolecule | Name: ADDomer: An engineered viral like particle derived from penton base protein of Human adenovirus B3 (Ad3). type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Recombinantly expressed and purified ADDomer complex (penton base dodecahedron). |
|---|---|
| Source (natural) | Organism:  Human adenovirus B3 Human adenovirus B3 |
| Molecular weight | Theoretical: 3.811834 MDa |
-Macromolecule #1: Penton protein
| Macromolecule | Name: Penton protein / type: protein_or_peptide / ID: 1 / Number of copies: 60 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human adenovirus B serotype 3 Human adenovirus B serotype 3 |
| Molecular weight | Theoretical: 63.598293 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MRRRAVLGGA VVYPEGPPPS YESVMQQQAA MIQPPLEAPF VPPRYLAPTE GRNSIRYSEL SPLYDTTKLY LVDNKSADIA SLNYQNDHS NFLTTVVQNN DFTPTEASTQ TINFDERSRW GGQLKTIMHT NMPNVNEYMF SNKFKARVMV SRKAPEGEFV T VNDGPVND ...String: MRRRAVLGGA VVYPEGPPPS YESVMQQQAA MIQPPLEAPF VPPRYLAPTE GRNSIRYSEL SPLYDTTKLY LVDNKSADIA SLNYQNDHS NFLTTVVQNN DFTPTEASTQ TINFDERSRW GGQLKTIMHT NMPNVNEYMF SNKFKARVMV SRKAPEGEFV T VNDGPVND TYDHKEDILK YEWFEFILPE GNFSATMTID LMNNAIIDNY LEIGRQNGVL ESDIGVKFDT RNFRLGWDPE TK LIMPGVY TYEAFHPDIV LLPGCGVDFT ESRLSNLLGI RKRHPFQEGF KIMYEDLEGG NIPALLDVTA YEESKKDTTT ARE TTTLAV AEETSEDVDD DITRGDTYIT ELEKQKREAA AAEVSRKKEL KIQPLEKDSK SRSYNVLEDK INTAYRSWYL SYNY GNPEK GIRSWTLLTT SDVTCGAEQV YWSLPDMMQD PVTFRSTRQV NNYPVVGAEL MPVFSKSFYN EQAVYSQQLR QATSL THVF NRFPENQILI RPPAPTITTV SENVPALTDH GTLPLRSSIR GVQRVTVTDA RRRTCPYVYK ALGIVAPRVL SSRTF UniProtKB: Penton protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 1060 / Average exposure time: 0.25 sec. / Average electron dose: 1.3125 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 130000 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Residue range: 61-542 / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | The model was adjusted to fit into the map manually using COOT before further iterative positional and B-factor refinement in real space using Phenix Real-Space refinement software. Final adjustments were carried out in COOT. |
| Refinement | Space: REAL / Protocol: BACKBONE TRACE |
| Output model |  PDB-6hcr: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)