+Search query
-Structure paper
| Title | Structural basis of sodium ion-dependent carnitine transport by OCTN2. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 17, Issue 1, Page 181, Year 2025 |
| Publish date | Nov 29, 2025 |
 Authors Authors | James S Davies / Yi C Zeng / Chelsea Briot / Simon H J Brown / Renae M Ryan / Alastair G Stewart /  |
| PubMed Abstract | Carnitine is essential for the import of long-chain fatty acids into mitochondria, where they are used for energy production. The carnitine transporter OCTN2 (novel organic cation transporter 2, ...Carnitine is essential for the import of long-chain fatty acids into mitochondria, where they are used for energy production. The carnitine transporter OCTN2 (novel organic cation transporter 2, SLC22A5) mediates carnitine uptake across the plasma membrane and as such facilitates fatty acid metabolism in most tissues. OCTN2 dysfunction causes systemic primary carnitine deficiency (SPCD), a potentially lethal disorder. Despite its importance in metabolism, the mechanism of high-affinity, sodium ion-dependent transport by OCTN2 is unclear. Here we report cryo-EM structures of human OCTN2 in three conformations: inward-facing ligand-free, occluded carnitine- and Na-bound, and inward-facing ipratropium-bound. These structures define key interactions responsible for carnitine transport and identify an allosterically coupled Na binding site housed within an aqueous cavity, separate from the carnitine-binding site. Combined with electrophysiology data, we provide a framework for understanding variants associated with SPCD and insight into how OCTN2 functions as the primary human carnitine transporter. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:41318751 / PubMed:41318751 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.72 - 3.06 Å |
| Structure data | EMDB-71540, PDB-9pdq: EMDB-71597, PDB-9pfb: EMDB-71735, PDB-9pmd: |
| Chemicals |  ChemComp-NA: 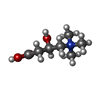 ChemComp-152:  ChemComp-NAG:  ChemComp-HOH: 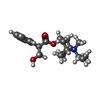 ChemComp-X8M: |
| Source |
|
 Keywords Keywords | TRANSPORT PROTEIN / transporter / carnitine transporter / organic cation transporter / SLC22 family / sodium-dependent transport / membrane protein / solute carrier / metabolic transport / fatty acid oxidation / OCTN2 / SLC22A5 / ipratropium / inhibitor |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers









 homo sapiens (human)
homo sapiens (human)