+Search query
-Structure paper
| Title | Structural insights into binding-site access and ligand recognition by human ABCB1. |
|---|---|
| Journal, issue, pages | EMBO J, Vol. 44, Issue 4, Page 991-991006, Year 2025 |
| Publish date | Jan 13, 2025 |
 Authors Authors | Devanshu Kurre / Phuoc X Dang / Le T M Le / Varun V Gadkari / Amer Alam /  |
| PubMed Abstract | ABCB1 is a broad-spectrum efflux pump central to cellular drug handling and multidrug resistance in humans. However, how it is able to recognize and transport a wide range of diverse substrates ...ABCB1 is a broad-spectrum efflux pump central to cellular drug handling and multidrug resistance in humans. However, how it is able to recognize and transport a wide range of diverse substrates remains poorly understood. Here we present cryo-EM structures of lipid-embedded human ABCB1 in conformationally distinct apo-, substrate-bound, inhibitor-bound, and nucleotide-trapped states at 3.4-3.9 Å resolution, in the absence of stabilizing antibodies or mutations. The substrate-binding site is located within one half of the molecule and, in the apo state, is obstructed by the transmembrane helix (TM) 4. Substrate and inhibitor binding are distinguished by major TM rearrangements and their ligand binding chemistry, with TM4 playing a central role in all conformational transitions. Furthermore, our data identify secondary structure-breaking residues that impart localized TM flexibility and asymmetry between the two transmembrane domains. The resulting structural changes and lipid interactions that are induced by substrate and inhibitor binding can predict substrate-binding profiles and may direct ABCB1 inhibitor design. |
 External links External links |  EMBO J / EMBO J /  PubMed:39806099 / PubMed:39806099 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.4 - 4.7 Å |
| Structure data | EMDB-45854, PDB-9cr8: EMDB-45903, PDB-9ctc: EMDB-45904, PDB-9ctf: EMDB-45906, PDB-9ctg:  EMDB-45931: HUMAN ABCB1 in SapNPs in complex with Taxol  EMDB-45932: HUMAN ABCB1 in SapNPs in complex with Zosuquidar |
| Chemicals |  ChemComp-UPL:  ChemComp-ATP: 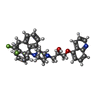 ChemComp-ZQU:  ChemComp-TA1:  ChemComp-MG:  ChemComp-AGS: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / TRANSLOCASE / ABCB1 / P-gp / apo form / nanoparticle / saposin A / MDR1 / p-glycoprotein / pgp / inhibitor bound / saposin / zosuquidar bound / substrate bound / taxol bound / php / ATPgammaS bound / non-hydrolysable nucleotide |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers











 homo sapiens (human)
homo sapiens (human)