+Search query
-Structure paper
| Title | Cryo-EM structures of wild-type and E138K/M184I mutant HIV-1 RT/DNA complexed with inhibitors doravirine and rilpivirine. |
|---|---|
| Journal, issue, pages | Proc Natl Acad Sci U S A, Vol. 119, Issue 30, Page e2203660119, Year 2022 |
| Publish date | Jul 26, 2022 |
 Authors Authors | Abhimanyu K Singh / Brent De Wijngaert / Marc Bijnens / Kris Uyttersprot / Hoai Nguyen / Sergio E Martinez / Dominique Schols / Piet Herdewijn / Christophe Pannecouque / Eddy Arnold / Kalyan Das /   |
| PubMed Abstract | Structures trapping a variety of functional and conformational states of HIV-1 reverse transcriptase (RT) have been determined by X-ray crystallography. These structures have played important roles ...Structures trapping a variety of functional and conformational states of HIV-1 reverse transcriptase (RT) have been determined by X-ray crystallography. These structures have played important roles in explaining the mechanisms of catalysis, inhibition, and drug resistance and in driving drug design. However, structures of several desired complexes of RT could not be obtained even after many crystallization or crystal soaking experiments. The ternary complexes of doravirine and rilpivirine with RT/DNA are such examples. Structural study of HIV-1 RT by single-particle cryo-electron microscopy (cryo-EM) has been challenging due to the enzyme's relatively smaller size and higher flexibility. We optimized a protocol for rapid structure determination of RT complexes by cryo-EM and determined six structures of wild-type and E138K/M184I mutant RT/DNA in complexes with the nonnucleoside inhibitors rilpivirine, doravirine, and nevirapine. RT/DNA/rilpivirine and RT/DNA/doravirine complexes have structural differences between them and differ from the typical conformation of nonnucleoside RT inhibitor (NNRTI)-bound RT/double-stranded DNA (dsDNA), RT/RNA-DNA, and RT/dsRNA complexes; the primer grip in RT/DNA/doravirine and the YMDD motif in RT/DNA/rilpivirine have large shifts. The DNA primer 3'-end in the doravirine-bound structure is positioned at the active site, but the complex is in a nonproductive state. In the mutant RT/DNA/rilpivirine structure, I184 is stacked with the DNA such that their relative positioning can influence rilpivirine in the pocket. Simultaneously, E138K mutation opens the NNRTI-binding pocket entrance, potentially contributing to a faster rate of rilpivirine dissociation by E138K/M184I mutant RT, as reported by an earlier kinetic study. These structural differences have implications for understanding molecular mechanisms of drug resistance and for drug design. |
 External links External links |  Proc Natl Acad Sci U S A / Proc Natl Acad Sci U S A /  PubMed:35858448 / PubMed:35858448 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.32 - 3.65 Å |
| Structure data | EMDB-14457, PDB-7z24: EMDB-14458, PDB-7z29: EMDB-14462, PDB-7z2d: EMDB-14463, PDB-7z2e: EMDB-14465, PDB-7z2g: EMDB-14466, PDB-7z2h: |
| Chemicals | 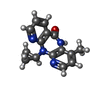 ChemComp-NVP:  ChemComp-T27: 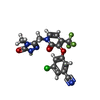 ChemComp-2KW: |
| Source |
|
 Keywords Keywords | TRANSFERASE / Reverse transcriptase / RT-aptamer complex / NON-NUCLEOSIDE INHIBITOR / P51 / P66 / Reverse transcriptase mutant / NNRTI resistance / NNRTI |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers















 human immunodeficiency virus type 1 bh10
human immunodeficiency virus type 1 bh10