+Search query
-Structure paper
| Title | Structural Basis for Binding of Potassium-Competitive Acid Blockers to the Gastric Proton Pump. |
|---|---|
| Journal, issue, pages | J Med Chem, Vol. 65, Issue 11, Page 7843-7853, Year 2022 |
| Publish date | Jun 9, 2022 |
 Authors Authors | Saki Tanaka / Mikio Morita / Tatsuya Yamagishi / Hridya Valia Madapally / Kenichi Hayashida / Himanshu Khandelia / Christoph Gerle / Hideki Shigematsu / Atsunori Oshima / Kazuhiro Abe /   |
| PubMed Abstract | As specific inhibitors of the gastric proton pump, responsible for gastric acidification, K-competitive acid blockers (P-CABs) have recently been utilized in the clinical treatment of gastric acid- ...As specific inhibitors of the gastric proton pump, responsible for gastric acidification, K-competitive acid blockers (P-CABs) have recently been utilized in the clinical treatment of gastric acid-related diseases in Asia. However, as these compounds have been developed based on phenotypic screening, their detailed binding poses are unknown. We show crystal and cryo-EM structures of the gastric proton pump in complex with four different P-CABs, tegoprazan, soraprazan, PF-03716556 and revaprazan, at resolutions reaching 2.8 Å. The structures describe molecular details of their interactions and are supported by functional analyses of mutations and molecular dynamics simulations. We reveal that revaprazan has a novel binding mode in which its tetrahydroisoquinoline moiety binds deep in the cation transport conduit. The mechanism of action of these P-CABs can now be evaluated at the molecular level, which will facilitate the rational development and improvement of currently available P-CABs to provide better treatment of acid-related gastrointestinal diseases. |
 External links External links |  J Med Chem / J Med Chem /  PubMed:35604136 PubMed:35604136 |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 2.76 - 3.5 Å |
| Structure data | EMDB-32299, PDB-7w4a:  PDB-7w47:  PDB-7w48:  PDB-7w49: |
| Chemicals |  ChemComp-MG:  ChemComp-RB: 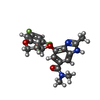 ChemComp-8BN:  ChemComp-NAG:  ChemComp-HOH:  ChemComp-CE1: 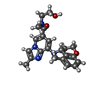 ChemComp-8BZ:  ChemComp-PCW: 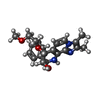 ChemComp-8CE: 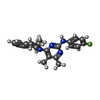 ChemComp-8CK:  ChemComp-NA: |
| Source |
|
 Keywords Keywords | HYDROLASE / Cation pump / P-type ATPase / gastric / proton pump / membrane protein / P-CAB |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers






