+Search query
-Structure paper
| Title | Structure and transport mechanism of the human cholesterol transporter ABCG1. |
|---|---|
| Journal, issue, pages | Cell Rep, Vol. 38, Issue 4, Page 110298, Year 2022 |
| Publish date | Jan 25, 2022 |
 Authors Authors | Da Xu / Yanyan Li / Fengrui Yang / Cai-Rong Sun / Jinheng Pan / Liang Wang / Zhi-Peng Chen / Shu-Cheng Fang / Xuebiao Yao / Wen-Tao Hou / Cong-Zhao Zhou / Yuxing Chen /  |
| PubMed Abstract | The reverse cholesterol transport pathway is responsible for the maintenance of human cholesterol homeostasis, an imbalance of which usually leads to atherosclerosis. As a key component of this ...The reverse cholesterol transport pathway is responsible for the maintenance of human cholesterol homeostasis, an imbalance of which usually leads to atherosclerosis. As a key component of this pathway, the ATP-binding cassette transporter ABCG1 forwards cellular cholesterol to the extracellular acceptor nascent high-density lipoprotein (HDL). Here, we report a 3.26-Å cryo-electron microscopy structure of cholesterol-bound ABCG1 in an inward-facing conformation, which represents a turnover condition upon ATP binding. Structural analyses combined with functional assays reveals that a cluster of conserved hydrophobic residues, in addition to two sphingomyelins, constitute a well-defined cholesterol-binding cavity. The exit of this cavity is closed by three pairs of conserved Phe residues, which constitute a hydrophobic path for the release of cholesterol in an acceptor concentration-dependent manner. Overall, we propose an ABCG1-driven cholesterol transport cycle initiated by sphingomyelin-assisted cholesterol recruitment and accomplished by the release of cholesterol to HDL. |
 External links External links |  Cell Rep / Cell Rep /  PubMed:35081353 PubMed:35081353 |
| Methods | EM (single particle) |
| Resolution | 3.26 Å |
| Structure data | EMDB-31547, PDB-7fdv: |
| Chemicals |  ChemComp-ATP: 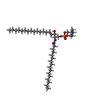 ChemComp-HWP:  ChemComp-CLR: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / Human ABCG1 / cholsterol transporter / ATP-bound / cholesterol |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers





 homo sapiens (human)
homo sapiens (human)