+Search query
-Structure paper
| Title | Structural mechanism of human TRPC3 and TRPC6 channel regulation by their intracellular calcium-binding sites. |
|---|---|
| Journal, issue, pages | Neuron, Vol. 110, Issue 6, Page 1023-11035.e5, Year 2022 |
| Publish date | Mar 16, 2022 |
 Authors Authors | Wenjun Guo / Qinglin Tang / Miao Wei / Yunlu Kang / Jing-Xiang Wu / Lei Chen /  |
| PubMed Abstract | TRPC3 and TRPC6 channels are calcium-permeable non-selective cation channels that are involved in many physiological processes. The gain-of-function (GOF) mutations of TRPC6 lead to familial focal ...TRPC3 and TRPC6 channels are calcium-permeable non-selective cation channels that are involved in many physiological processes. The gain-of-function (GOF) mutations of TRPC6 lead to familial focal segmental glomerulosclerosis (FSGS) in humans, but their pathogenic mechanism remains elusive. Here, we report the cryo-EM structures of human TRPC3 in both high-calcium and low-calcium conditions. Based on these structures and accompanying electrophysiological studies, we identified both inhibitory and activating calcium-binding sites in TRPC3 that couple intracellular calcium concentrations to the basal channel activity. These calcium sensors are also structurally and functionally conserved in TRPC6. We uncovered that the GOF mutations of TRPC6 activate the channel by allosterically abolishing the inhibitory effects of intracellular calcium. Furthermore, structures of human TRPC6 in complex with two chemically distinct inhibitors bound at different ligand-binding pockets reveal different conformations of the transmembrane domain, providing templates for further structure-based drug design targeting TRPC6-related diseases such as FSGS. |
 External links External links |  Neuron / Neuron /  PubMed:35051376 PubMed:35051376 |
| Methods | EM (single particle) |
| Resolution | 2.7 - 3.9 Å |
| Structure data | EMDB-30903, PDB-7dxb: EMDB-30904, PDB-7dxc: EMDB-30905, PDB-7dxd: EMDB-30906, PDB-7dxe: EMDB-30907, PDB-7dxf: EMDB-30908, PDB-7dxg: |
| Chemicals |  ChemComp-POV:  ChemComp-Y01:  ChemComp-ZN:  ChemComp-CA:  ChemComp-98R: 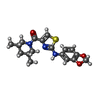 ChemComp-W99:  ChemComp-HOR: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / TRPC3 / TRPC / calcium / channel / gain of function / TRPC6 / BTDM / FSGS / SAR7334 |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers















 homo sapiens (human)
homo sapiens (human)