+Search query
-Structure paper
| Title | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E receptor EP2 subtype. |
|---|---|
| Journal, issue, pages | Sci Adv, Vol. 7, Issue 14, Year 2021 |
| Publish date | Apr 2, 2021 |
 Authors Authors | Changxiu Qu / Chunyou Mao / Peng Xiao / Qingya Shen / Ya-Ni Zhong / Fan Yang / Dan-Dan Shen / Xiaona Tao / Huibing Zhang / Xu Yan / Ru-Jia Zhao / Junyan He / Ying Guan / Chao Zhang / Guihua Hou / Peng-Ju Zhang / Guige Hou / Zijian Li / Xiao Yu / Ren-Jie Chai / You-Fei Guan / Jin-Peng Sun / Yan Zhang /  |
| PubMed Abstract | Selective modulation of the heterotrimeric G protein α S subunit-coupled prostaglandin E (PGE) receptor EP2 subtype is a promising therapeutic strategy for osteoporosis, ocular hypertension, ...Selective modulation of the heterotrimeric G protein α S subunit-coupled prostaglandin E (PGE) receptor EP2 subtype is a promising therapeutic strategy for osteoporosis, ocular hypertension, neurodegenerative diseases, and cardiovascular disorders. Here, we report the cryo-electron microscopy structure of the EP2-G complex with its endogenous agonist PGE and two synthesized agonists, taprenepag and evatanepag (CP-533536). These structures revealed distinct features of EP2 within the EP receptor family in terms of its unconventional receptor activation and G protein coupling mechanisms, including activation in the absence of a typical W "toggle switch" and coupling to G via helix 8. Moreover, inspection of the agonist-bound EP2 structures uncovered key motifs governing ligand selectivity. Our study provides important knowledge for agonist recognition and activation mechanisms of EP2 and will facilitate the rational design of drugs targeting the PGE signaling system. |
 External links External links |  Sci Adv / Sci Adv /  PubMed:33811074 / PubMed:33811074 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.8 - 2.9 Å |
| Structure data | EMDB-30489, PDB-7cx2: EMDB-30490, PDB-7cx3: EMDB-30491, PDB-7cx4: |
| Chemicals | 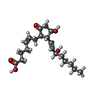 ChemComp-P2E:  ChemComp-HOH: 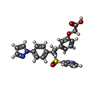 ChemComp-GNO: 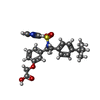 ChemComp-GM9: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / GPCR / EP2 / Complex / PGE2 / Taprenepag / Evatanepag |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers









 homo sapiens (human)
homo sapiens (human)