+Search query
-Structure paper
| Title | Structures of the human dopamine D3 receptor-G complexes. |
|---|---|
| Journal, issue, pages | Mol Cell, Vol. 81, Issue 6, Page 1147-11159.e4, Year 2021 |
| Publish date | Mar 18, 2021 |
 Authors Authors | Peiyu Xu / Sijie Huang / Chunyou Mao / Brian E Krumm / X Edward Zhou / Yangxia Tan / Xi-Ping Huang / Yongfeng Liu / Dan-Dan Shen / Yi Jiang / Xuekui Yu / Hualiang Jiang / Karsten Melcher / Bryan L Roth / Xi Cheng / Yan Zhang / H Eric Xu /   |
| PubMed Abstract | The dopamine system, including five dopamine receptors (D1R-D5R), plays essential roles in the central nervous system (CNS), and ligands that activate dopamine receptors have been used to treat many ...The dopamine system, including five dopamine receptors (D1R-D5R), plays essential roles in the central nervous system (CNS), and ligands that activate dopamine receptors have been used to treat many neuropsychiatric disorders. Here, we report two cryo-EM structures of human D3R in complex with an inhibitory G protein and bound to the D3R-selective agonists PD128907 and pramipexole, the latter of which is used to treat patients with Parkinson's disease. The structures reveal agonist binding modes distinct from the antagonist-bound D3R structure and conformational signatures for ligand-induced receptor activation. Mutagenesis and homology modeling illuminate determinants of ligand specificity across dopamine receptors and the mechanisms for G protein coupling. Collectively our work reveals the basis of agonist binding and ligand-induced receptor activation and provides structural templates for designing specific ligands to treat CNS diseases targeting the dopaminergic system. |
 External links External links |  Mol Cell / Mol Cell /  PubMed:33548201 PubMed:33548201 |
| Methods | EM (single particle) |
| Resolution | 2.7 - 3.0 Å |
| Structure data | EMDB-30410, PDB-7cmu: EMDB-30411, PDB-7cmv: |
| Chemicals | 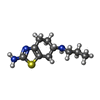 ChemComp-G6L: 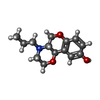 ChemComp-G6O: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / G protein-coupled receptor / Dopamine receptor |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers







 homo sapiens (human)
homo sapiens (human)

